-
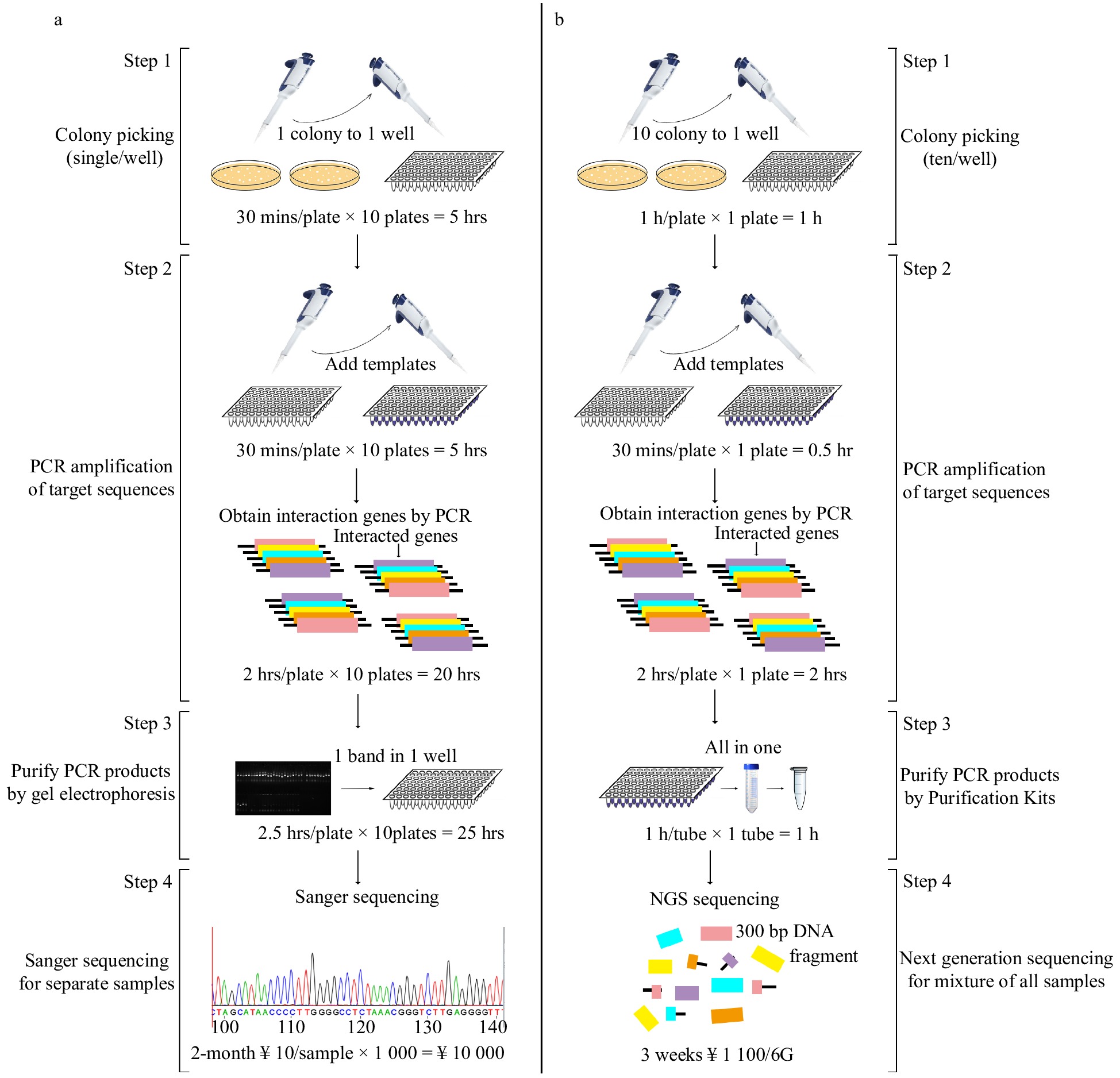
Figure 1. Flowchart of traditional Y2H and Y2H-seq. (a) Traditional Y2H, (b) Y2H-seq technology.
-

Figure 2. Y2H verification of PtDAL1 interacting proteins. Yeast liquid dilution concentration: 100, 10−2, 10−3, 10−4 (from left to right).
-
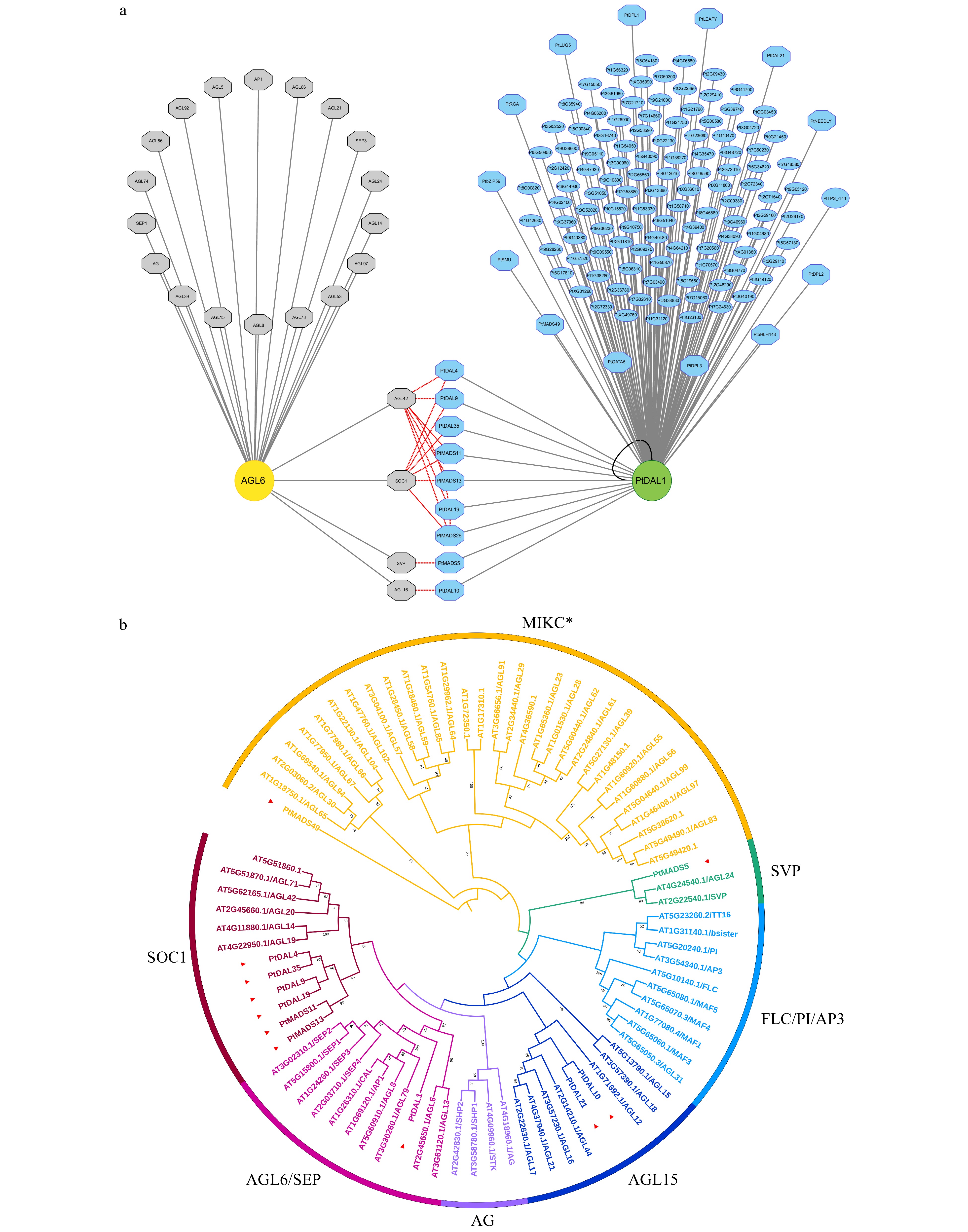
Figure 3. The interactome of PtDAL1 and its Arabidopsis homologue AtAGL6. (a) Gray, AtAGL6 interacted proteins in Arabidopsis. Blue, PtDAL1 interacted proteins in P. tabuliformis. Red lines indicate the homologs between Arabidopsis and P. tabuliformis. Black lines indicate homologous dimer. Octagon, transcription factors. (b) The phylogenetic analysis of DIPs in MADS-box gene family. Red inverted triangle, DIPs of MADS-box family in P. tabuliformis.
-
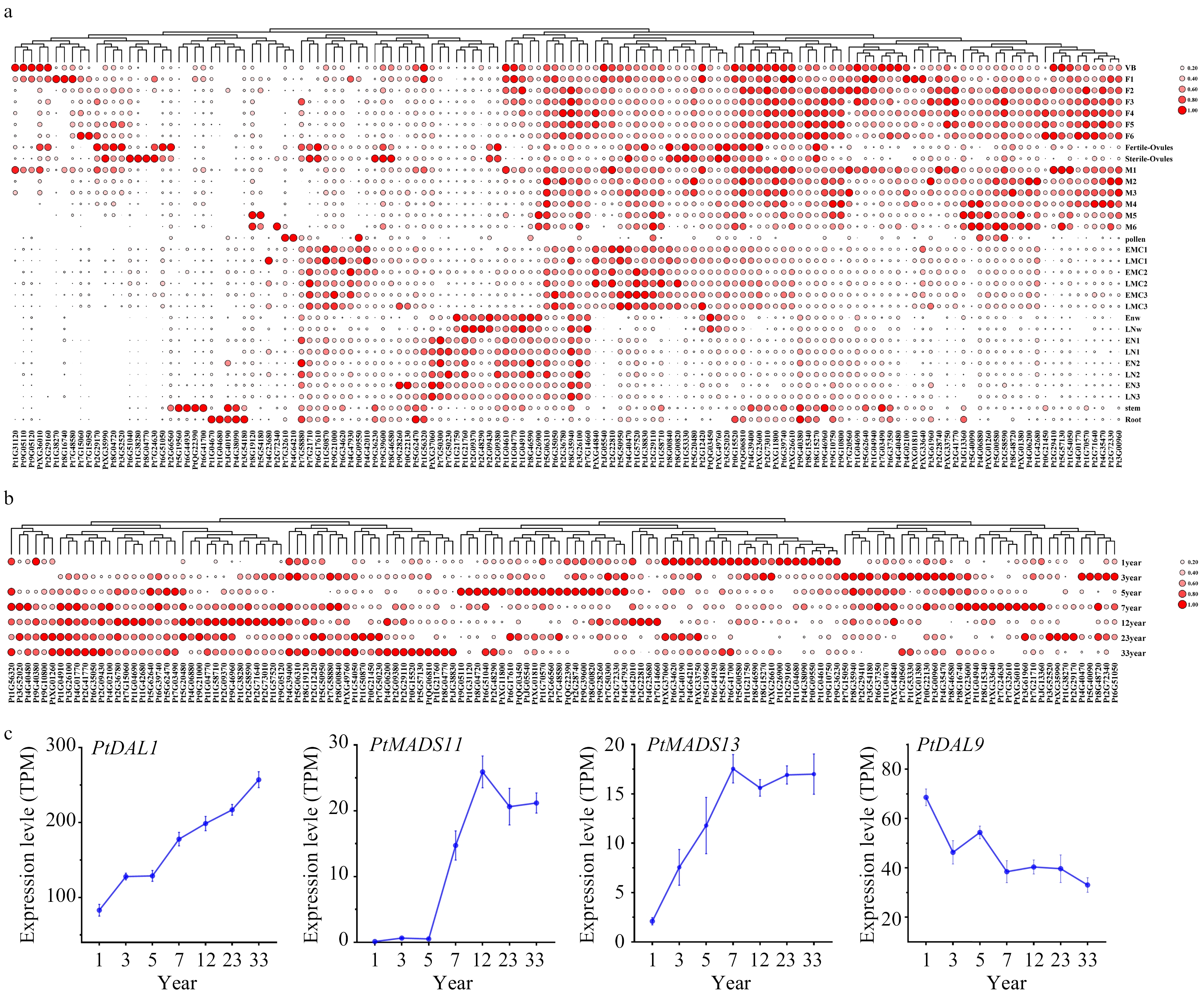
Figure 4. The expression profiles of PtDAL1 interacting proteins. (a) Expression patterns of PtDAL1 interacting proteins in different tissues, (b) expression patterns of PtDAL1 interacting proteins at different ages, (c) expression patterns of PtDAL1, PtDAL9, PtMADS11 and PtMADS13 at different ages.
-
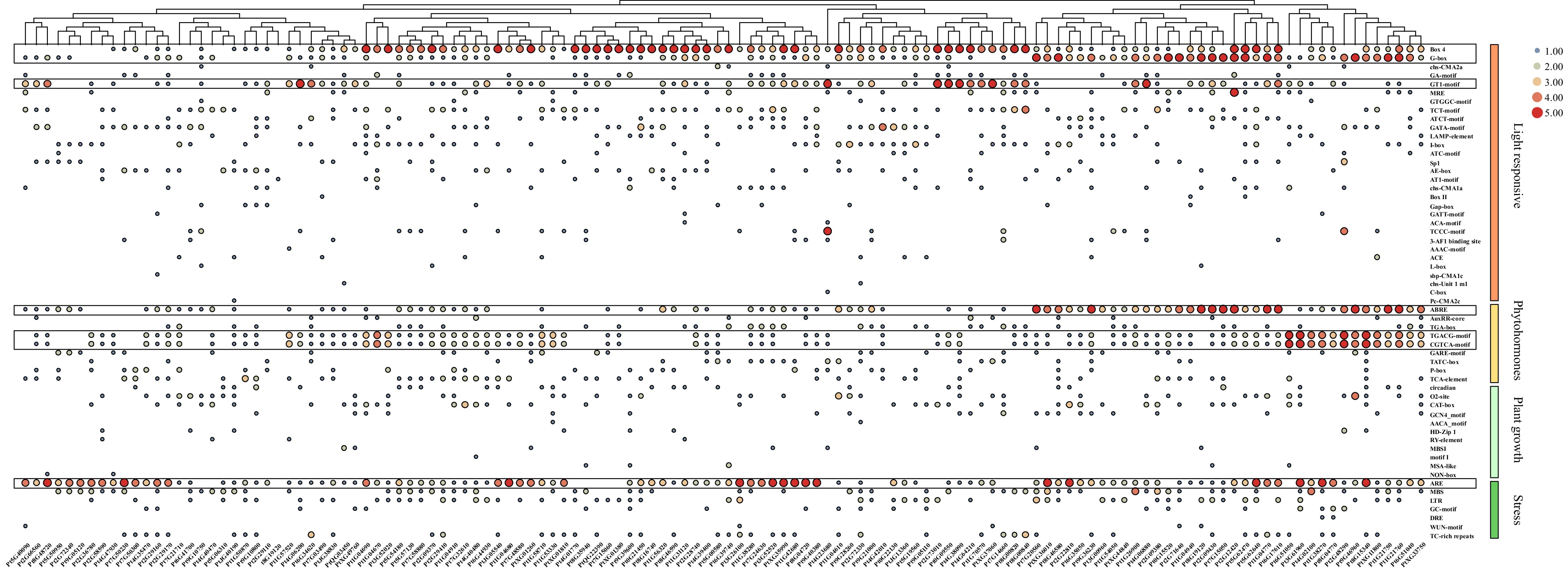
Figure 5. Cis-acting element analysis of PtDAL1 interacting proteins. ABRE, ABA responsive motif; TGACG motif and CGTCA motif, MeJA responsive motif.
-
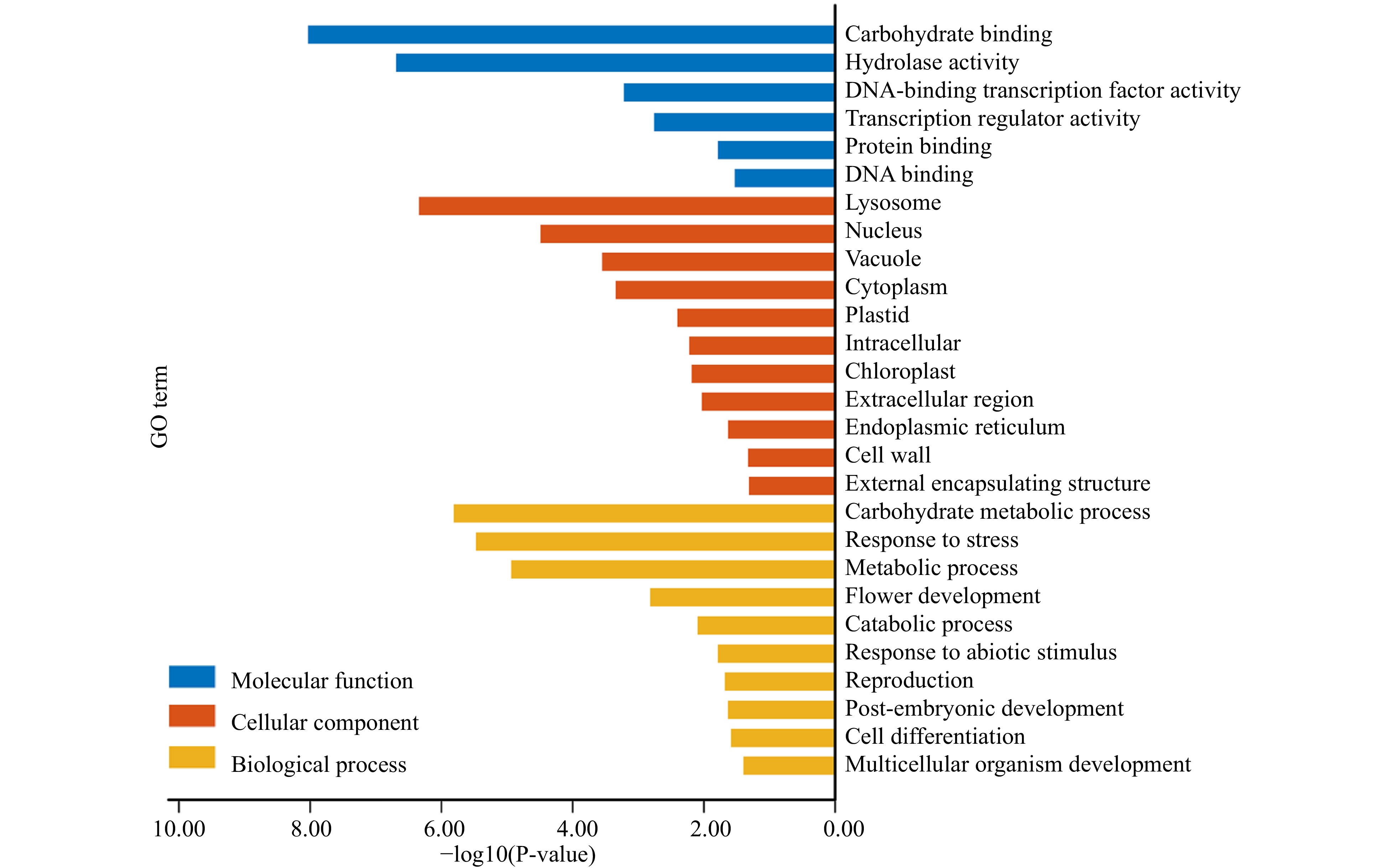
Figure 6. GO functions of PtDAL1 interacted proteins.
-
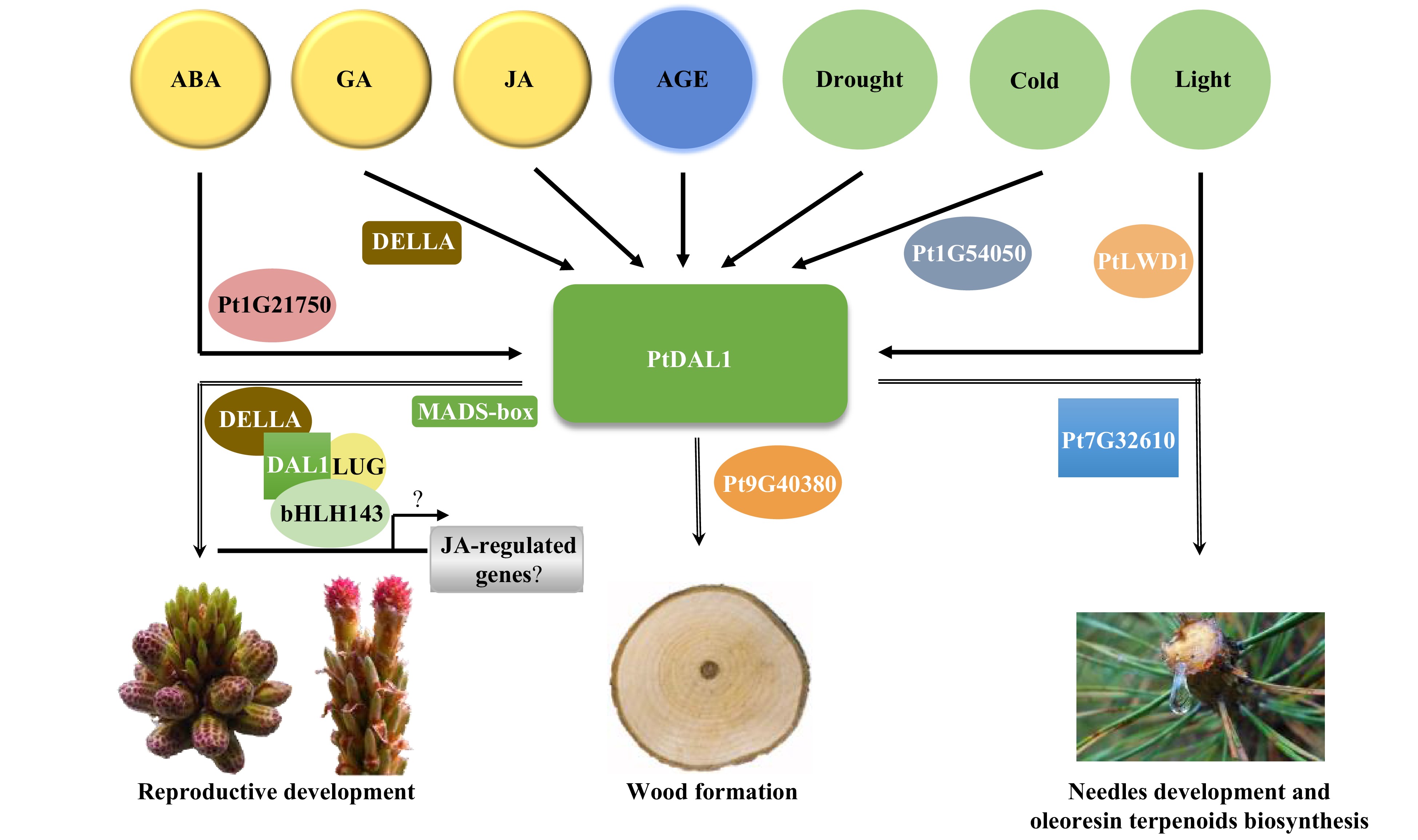
Figure 7. A predictive model for the PtDAL1-mediated growth and development regulation in P. tabuliformis.
-
Gene_ID\Symbol At_homo e-value At_gene_symbol TF/TR/PK EST count Pt0G09550 AT1G74310 1.32E-39 AtHSP101 33,639 Pt0G15520 AT4G38510 0 AtVAB2 34,522 Pt0G21450 AT5G08640 2.8E-125 AtFLS1 28,899 Pt0G22130 AT4G37480 6.7E-137 22 Pt1G04610\PtDAL9 AT2G45660 3.41E-65 AGAMOUS-LIKE 20 (AtSOC1) TF 66,859 Pt1G04670\PtDAL19 AT2G45660 8.53E-57 AGAMOUS-LIKE 20 (AtSOC1) TF 573 Pt1G04680 − 31 Pt1G04770\PtMADS11 AT2G45660 2.34E-53 AGAMOUS-LIKE 20 (AtSOC1) TF 688 Pt1G04910\PtMADS13 AT2G45660 7.91E-61 AGAMOUS-LIKE 20 (AtSOC1) TF 257 Pt1G04940\PtMADS26 AT2G45660 1.37E-61 AGAMOUS-LIKE 20 (AtSOC1) TF 78 Pt1G21750 AT5G42270 0 VARIEGATED 1 (VAR1) 116,184 Pt1G21760 AT5G42270 0 VARIEGATED 1 (VAR1) 40 Pt1G26900 AT1G01090 0 PDH-E1 ALPHA 330 Pt1G31120 AT2G05790 5.67E-74 57 Pt1G38270 AT2G05790 1.95E-81 13,837 Pt1G38280 AT2G05790 1.95E-81 7,453 Pt1G42680 AT5G63480 1.18E-37 MEDIATOR 30 (MED30) TR 210,880 Pt1G50870 AT5G55160 6.87E-45 AtSUMO2 26 Pt1G53330 AT4G31200 1E-178 156,474 Pt1G54050 AT2G21660 8.22E-40 GLYCINE RICH PROTEIN 7 (AtGRP7) 11 Pt1G56320 − 52 Pt1G57520 AT3G05530 0 RPT5A 201,789 Pt1G58710 AT5G55160 5.72E-33 AtSUMO2 48,291 Pt1G70570 − 13 Pt2G09370 AT1G09070 4.32E-36 AtSRC2 18,394 Pt2G09380 − 11,074 Pt2G09430 − 62,709 Pt2G12420 AT2G46210 0 AtSLD2 388 Pt2G22810\PtLEAFY AT5G61850 2.7E-115 LEAFY 3 (LFY3) TF 10 Pt2G28740\PtbHLH143 AT1G59640 1.27E-38 BIG PETAL (BPE); ZCW32 TF 97 Pt2G29110 AT5G14040 0 MPT3 52,616 Pt2G29160 AT5G55180 6.73E-42 30 Pt2G29170 − 36 Pt2G29410 − 133 Pt2G36780 AT3G09970 3.2E-138 RHIZOBIALE-LIKE PHOSPHATASE 2 (RLPH2) 72 Pt2G41770\PtDAL10 AT1G26310 4.84E-47 CAULIFLOWER (CAL) TF 4 Pt2G48290 − 10 Pt2G58590 AT1G63000 0 UER1 56,441 Pt2G66560 − 16,151 Pt2G71640 AT1G75290 2.28E-97 42,597 Pt2G72330 − 669 Pt2G72340 − 33 Pt2G73010 − 16,500 Pt3G00960 − 16 Pt3G26100 AT2G29630 0 THIAMINC (THIC) 136,084 Pt3G52020 AT1G47710 2.7E-133 AtSERPIN1 853 Pt3G52520 − 30 Pt3G54180\PtMADS49 TF 1 Pt3G61960 AT4G32460 9.17E-63 AtHA1-1 119 Pt4G01770\PtTPS_di41 AT4G16730 1.72E-72 TERPENE SYNTHASE 02 (TPS02) 25 Pt4G02100 − 91 Pt4G06200 − 551,090 Pt4G06880 AT4G32470 4.21E-52 2,220,400 Pt4G23680 − 575 Pt4G35470 AT5G21060 2.6E-50 554 Pt4G38090 − 31 Pt4G39400 AT5G09760 1.6E-155 117 Pt4G40470 AT3G05530 1.46E-88 RPT5A 21,709 Pt4G40480 AT3G05530 3.5E-127 RPT5A 21,180 Pt4G42010 AT1G74310 9.99E-92 AtHSP101 1,847 Pt4G47930 − 49 Pt4G64210 AT2G04160 4.49E-46 AUXIN-INDUCED IN ROOT CULTURES 3 (AIR3) 10 Pt5G00580 AT1G21980 4E-146 AtPIP5K1; AtPIPK1 PK 98,948 Pt5G06310 AT3G46740 0 TOC75-III; MAR1 349 Pt5G19560 AT1G75030 2.04E-95 AtLP-3 17,623 Pt5G20480\PtGATA5 TF 1 Pt5G40090 − 12 Pt5G50950 AT3G05530 5.4E-143 RPT5A 85,346 Pt5G54180 AT5G43060 0 ESPONSIVE TO DEHYDRATION 21B (RD21B) 27 Pt5G57130 AT5G43060 5.3E-131 ESPONSIVE TO DEHYDRATION 21B (RD21B) 41 Pt5G62470\PtDAL4 AT2G45660 1.96E-67 AGAMOUS-LIKE 20 (AtSOC1) TF 1,003,270 Pt5G62640\PtDAL35 AT2G45660 2.36E-67 AGAMOUS-LIKE 20 (AtSOC1) TF 182,559 Pt6G17610 AT2G21660 7.65E-43 AtGRP7 2,751 Pt6G34620 AT2G21660 7.48E-41 AtGRP7 518 Pt6G35050\PtDAL1 AT2G45650 1.44E-81 AGAMOUS-LIKE 6 (AGL6) TF 126 Pt6G37350\PtMADS5 AT2G22540 3.69E-59 SHORT VEGETATIVE PHASE (SVP) TF 9 Pt6G39740 AT3G07320 8.7E-174 290,696 Pt6G41700 AT4G25200 3.8E-36 ATHSP23.6-MITO 157 Pt6G44930 − 23 Pt6G51040 AT4G26830 7E-145 202,392 Pt6G51050 AT2G05790 0 4,185,410 Pt7G03490 AT3G04880 2.3E-137 DNA-DAMAGE-REPAIR/TOLERATION 2 (DRT102) 224 Pt7G14660 AT5G42800 9.3E-164 DFR 2,048 Pt7G15050 AT5G06570 1.48E-37 21,889 Pt7G15060 AT1G68620 3.89E-50 73,081 Pt7G20560 AT3G23900 4.2E-120 IRR 4,284 Pt7G21710 − 6,227 Pt7G24630 − 14 Pt7G32610 AT3G05500 3.24E-70 SRP3; LDAP3 12,253 Pt7G48580 AT5G55180 1.85E-85 53 Pt7G50230 AT3G45310 1.3E-174 32 Pt7G50300 AT3G45310 2E-174 4,816 Pt7G58880 AT5G43060 9.3E-122 RD21B 8,270 Pt8G00820 AT1G18250 4.7E-107 ATLP-1 7,596 Pt8G00840 AT1G18250 4.5E-107 ATLP-1 94,972 Pt8G04720 AT5G55180 8.1E-82 135 Pt8G04770 AT5G55180 1.27E-88 5,885 Pt8G15270\PtDPL1 AT1G14920 6.5E-170 RESTORATION ON GROWTH ON AMMONIA 2 (RGA2) TF 9 Pt8G15340\PtDPL2 AT2G01570 1.1E-128 REPRESSOR OF GA1-3 1 (RGA1) TF 64 Pt8G16740 − 27 Pt8G19120 AT5G46290 0 3-KASI 101 Pt8G35940 AT5G47000 4.2E-125 75,498 Pt8G46580 AT5G43060 0 RD21B 14,683 Pt8G46590 AT5G43060 0 RD21B 186,050 Pt8G48720 AT3G05530 1.5E-143 RPT5A 13,449 Pt9G05110 − 415 Pt9G05120 − 28 Pt9G10750 − 11,338 Pt9G10800 − 8,111 Pt9G21000 AT5G38660 1.37E-61 APE1 87,565 Pt9G28260 − 372 Pt9G36230 AT4G23160 1.7E-116 CYSTEINE-RICH RLK 8 (CRK8) PK 17 Pt9G39600 AT4G12070 1.1E-151 95,337 Pt9G40380 AT1G55000 4.51E-87 26,718 Pt9G46960 AT2G17420 0 NTR2; AtNTRA 61,919 PtJG05540\PtNEEDLY AT5G61850 1.22E-88 LEAFY 3 (LFY3) TF 24 PtJG13360 AT1G54320 7.1E-173 12 PtJG38830 AT1G24510 0 CCT5 30,744 PtJG40190 AT3G04880 1.16E-53 DRT102 271 PtQG03450/PtLWD1 AT1G12910 0 AtAN11 41,358 PtQG06810\PtSMU AT2G26460 2.8E-113 SUPPRESSORS OF MEC-8 AND UNC-52 2 (SMU2) 1 PtQG22390 AT4G27670 1.21E-64 HEAT SHOCK PROTEIN 21 (HSP21) 22,925 PtXG01260 AT1G18640 2.4E-115 PSP1 66,835 PtXG01380 AT1G18640 1.6E-115 PHOSPHOSERINE PHOSPHATASE 1 (PSP1) 49 PtXG01810 AT5G21060 1.92E-53 30 PtXG11800 − 43 PtXG23600\PtbZIP59 AT4G38900 1.05E-89 BASIC LEUCINE-ZIPPER 29 (BZIP29) TF 1 PtXG26610\PtLUG5 AT4G32551 0 LEUNIG (LUG) TR 7 PtXG33640\PtRGA AT2G01570 2.2E-172 REPRESSOR OF GA1-3 1 (RGA1) TF 5 PtXG33750\PtDPL3 AT2G01570 2.2E-172 REPRESSOR OF GA1-3 1 (RGA1)) TF 133 PtXG35990 − 56 PtXG36010 AT4G26830 6.01E-90 37 PtXG37060 AT3G44620 3.69E-86 106,593 PtXG44840\PtDAL21 AT1G24260 4.83E-48 AGAMOUS-LIKE 9 (AGL9); SEPALLATA3 (SEP3) TF 13 PtXG49760 AT4G29040 0 RPT2a 92,990 Table 1. PtDAL1 interacting proteins annotation.
Figures
(7)
Tables
(1)