-
Figure 1. I. latifolia and 'Kudingcha'. (a) I. latifolia tree, (b) two types of 'Kudingcha' processed using young and old leaves.
-

Figure 2. Amino acid sequence alignment of I. latifolia KS (IlKS) with representative KSs from other plants, including Scoparia dulcis KS (SdKS), Arabidopsis thaliana KS (AtKS), Oryza sativa KS (OsKS), Populus trichocarpa KS (PtTPS19). The two conserved domains (DDxxD, NSE/DTE) binding to the diphosphate moiety of the substrate by magnesium ions are indicated using red lines. Conserved sequence elements are highlighted, and the shading indicates the conservation levels.
-
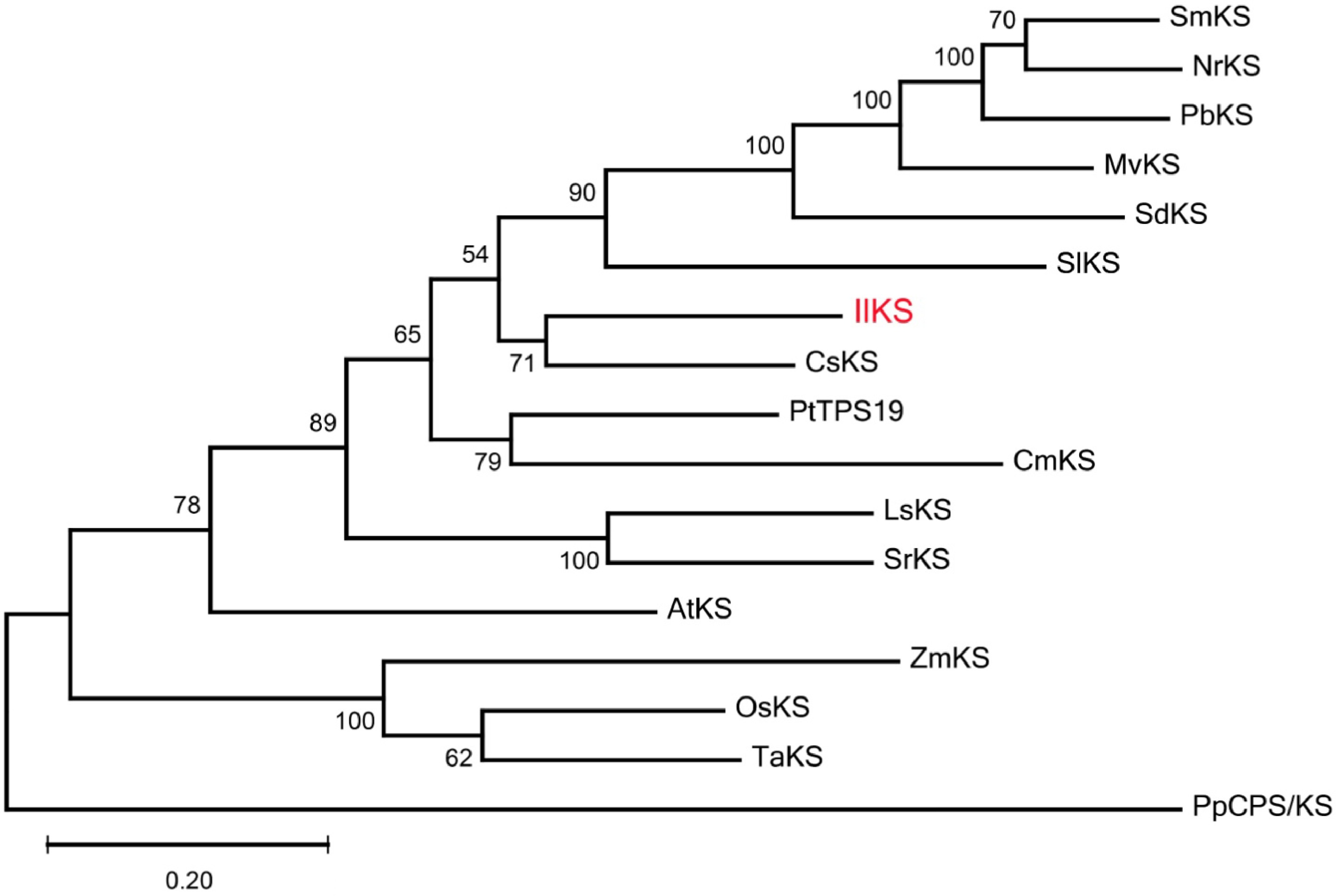
Figure 3. Phylogenetic tree of functionally characterized ent-kaurene synthases (KSs). The Maximum likelihood method was used to construct the tree with 500 bootstrap repetitions. Ent-kaurene synthase from I. latifolia (IlKS) is marked in red. The KSs from other plants used in the phylogenetic analysis are shown in Supplemental Table S2.
-
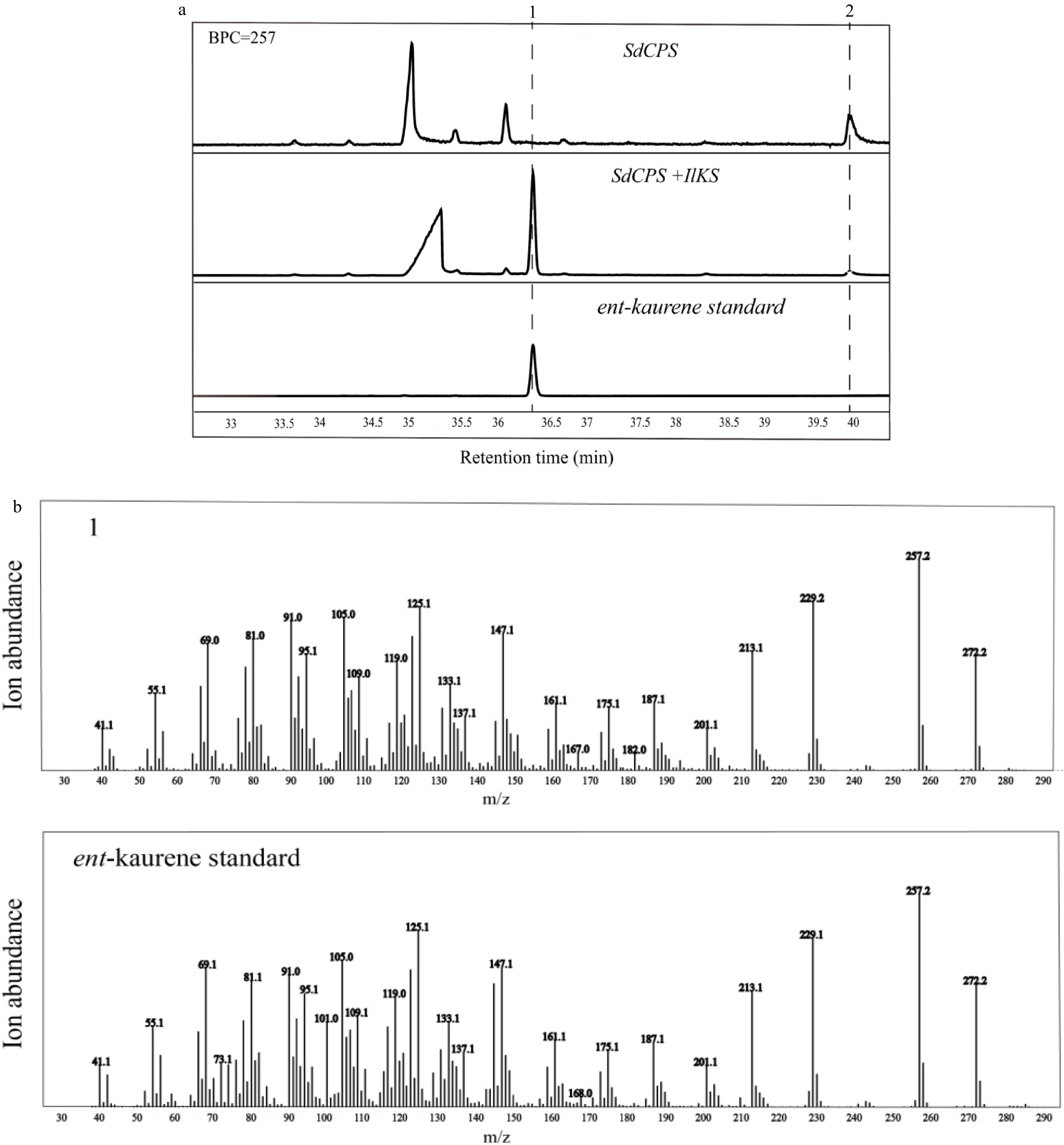
Figure 4. GC-MS analysis of diterpene products from hexane extracts of recombinant bacteria. (a) Selective ion chromatograms; (b) Mass spectra; 1, ent-kaurene; 2, ent-copalol.
-
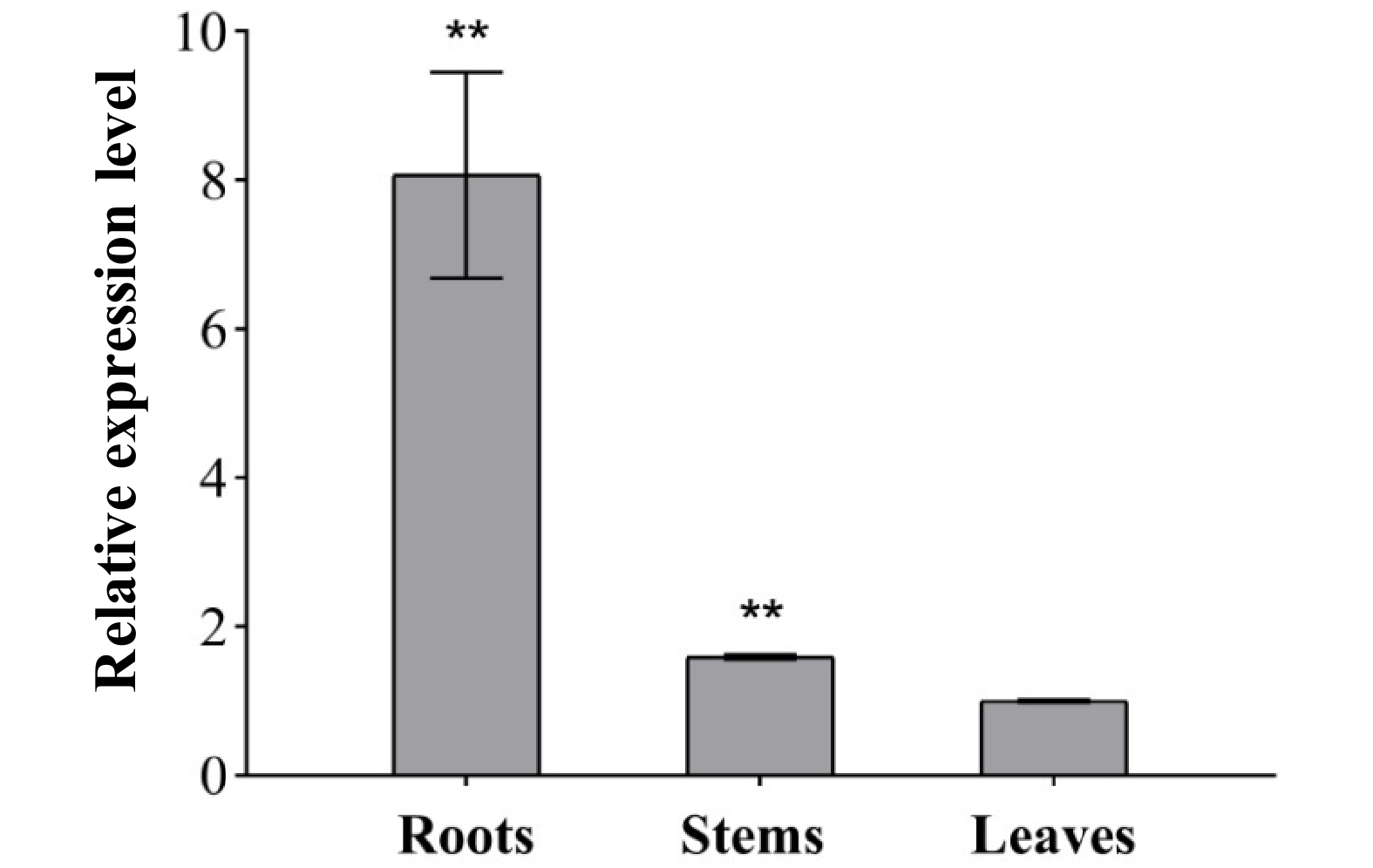
Figure 5. Relative transcript abundance of IlKS in roots, stems and leaves of I. latifolia. Transcript abundance was measured using qRT-PCR and normalized to the internal reference actin. The 2−ΔΔCT method was used to calculate the relative values. Asterisks indicate significant differences compared with the expression level in leaves. (** p < 0.01).
Figures
(5)
Tables
(0)