-
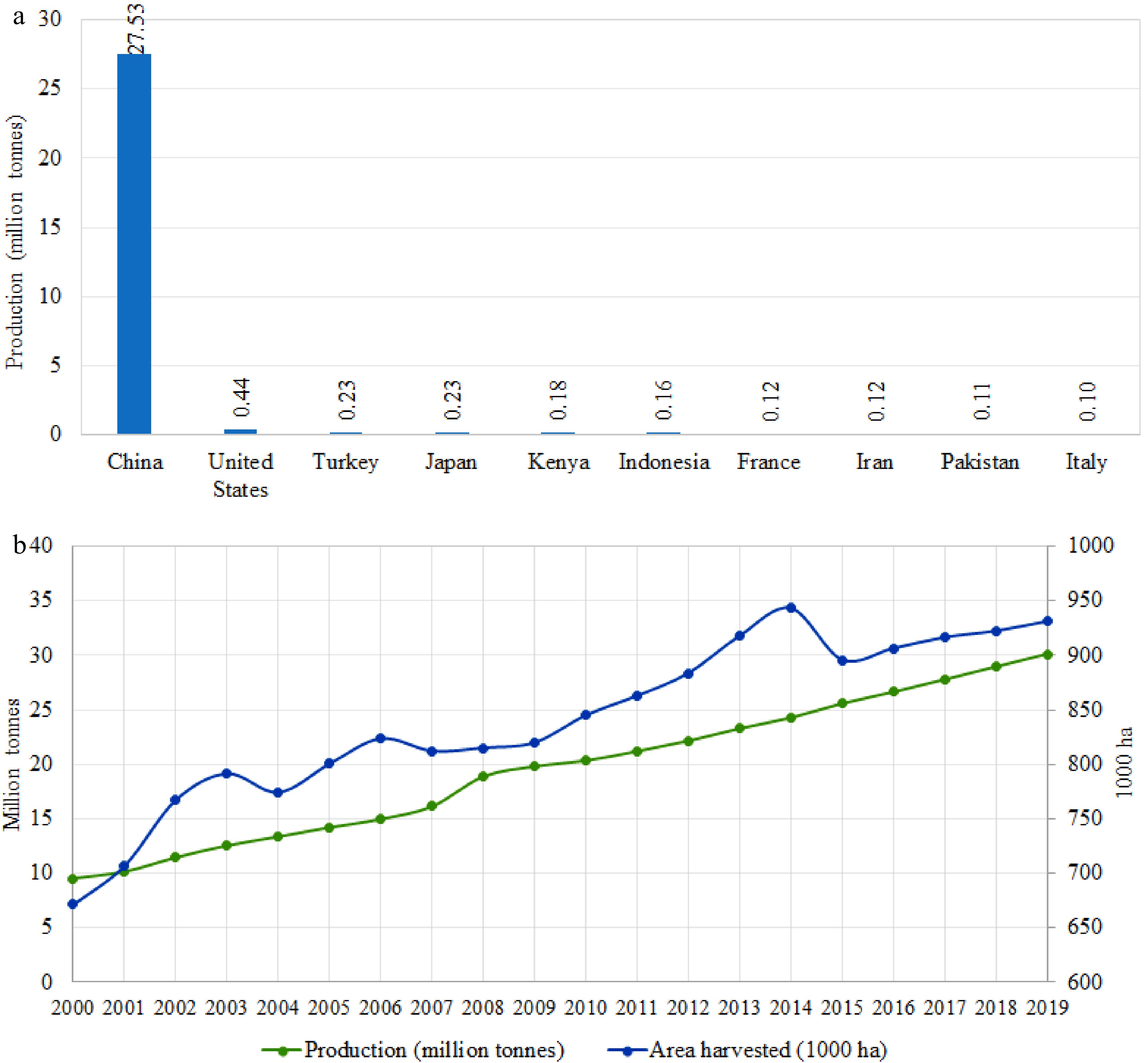
Figure 1. Worldwide spinach production. (a) The top ten spinach producing countries. (b) Global spinach production and harvest area in the past two decades.
-

Figure 2. Female (left) and male (right) spinach plants. Spinach plants are kept in isolated crossing blocks to generate seeds (center).
-

Figure 3. An overview of genome enabled breeding prospects of spinach.
-

Figure 4. High density spinach planting in Salinas, California and Yuma, Arizona, USA.
-

Figure 5. Signs and symptoms of downy mildew disease in spinach. Plants (middle two) inoculated with P. effusa in the greenhouse show sporulation and chlorosis. Baby leaf spinach plants growing in commercial field conditions, the resistant cultivars (left) are clean while the susceptible cultivars (right) are entirely infected.
-
Resources generated Outcomes of the activities References DNA markers 12 nuclear SSR markers were identified and amplified in spinach. [53] Genetic map 110 markers (101 AFLP and 9 SSR markers) were assigned to linkage groups. The gender determination gene was mapped to the linkage group 3 at 101.5 cM nearby five other markers between 97.4−102.6 cM. [17] DNA markers Target region amplification polymorphism (TRAP) markers were generated and were used to evaluate the genetic diversity among spinach germplasm accessions and commercial cultivars. [44] Bacterial artificial chromosome (BAC) library construction BAC library was constructed from a near-isogenic line (NIL1). Fourteen sequences analogous to known plant disease resistance genes were identified. Of the 14, nine contained nucleotide binding sites leucine-rich-repeat (NBS-LRR) domains. One hundred SSR loci were identified. One primers pair designed from these putative resistance genes was closely linked to the RPF1 locus and cosegregated with the DM1 marker. [54] DNA markers SSR loci were mined from Illumina sequences and a set of 85 polymorphic SSR markers were validated in 48 spinach accessions. [55] DNA markers Thirtyfour new polymorphic SSR markers were identified and genetic diversity was assessed on Chinese spinach accessions. [56] DNA markers Around 6,000 polymorphic SSRs reported in spinach following in silico genome sequence comparison. [35] Whole genome sequence A draft genome sequence of spinach cultivar was assembled to 498 Mb. [57] Annotation and gene set development Genome wide gene set was developed for spinach using the draft genome assembly[57] and mRNA sequencing from inbred spinach cultivar Viroflay. [58] Transcriptome sequences Transcriptomes from nine Spinacia accessions (three each from cultivated S. oleracea, and two wild S. turkestanica, and S. tetrandra) were sequenced at a high depth, genes were functionally annotated, Gene Ontology (GO) terms were assigned, and metabolic pathways were predicted. The SNP variants detected from the transcriptome assembly among the sequenced accessions were used to infer phylogeny, evolution, and domestication history. [46] Whole genome assembly (Spov1) The Spov1 genome sequence assembly developed for an inbred spinach line Sp75 provides a comprehensive genomic resource for spinach. The genome sequences were assembled to six linkage group that covers 463.4 Mb constituting 47% of the assembled genome. A total of 25,495 protein-coding genes were predicted, of which 139 NBS-LRR genes were identified that are known to provide disease resistance in plants. [30] Transcriptome assembly panel The transcriptome sequencing of 120 cultivated and wild Spinacia accessions resulted in a large number of transcribed variants and gene expression profiles. Nucleotide diversity and selection sweeps analysis were performed. The S. tetrandra was highly diverse compared to the other two Spinacia species, and S. turkestanica was the progenitor of cultivated spinach S. oleracea. [30] Spinach genomic database A publicly accessible database storing the reference spinach genome sequence, predicted functional annotations, gene expression profiles, and genetic variations based on transcriptome sequences of 120 cultivated and wild Spinacia accessions, and a platform to query and analyze genomic data were developed and hosted at SpinachBase (www.spinachbase.org). [60] Spinach genome assembly (Spov3) Another comprehensive whole genome assembly Spov3 generated for monoecious cultivar Viroflay (https://phytozome-next.jgi.doe.gov/info/Soleracea_Spov3). The genome was assembled to 913.5 Mb and the six main pseudomolecules comprised 745 Mb (81.6% of the genome) and predicted 34,877 genes in spinach. [61] Spinach genome assembly (SOL_r1.1) The SOL_r1.1 genome of breeding line 03-009 was assembled to 935.7 Mb of which 686.6 Mb (73.5%) was anchored to six chromosomes (http://spinach.kazusa.or.jp/index.html). [62] Spinach genome assembly (Monoe-Viroflay) Chromosome-scale reference genome assembly comprising 894.3 Mb with 879.2 Mb (98.32%) anchored to six chromosomes, and 28, 964 protein-coding genes were predicted. Genome resequencing of 305 spinach accessions and genome wide variant data are available. [34] Whole genome resequenced (WGR) GWAS panel A large set of 480 USDA accessions and commercial cultivars have been sequenced at a lower coverage to serve as the association panel for spinach. Several economically important traits have been phenotyped and genome wide association analysis (GWAS) are conducted to map the trait at a high-resolution mapping to identify causal variants. [63] Table 1. Summary of major genetic and genomic resources available for spinach.
Figures
(5)
Tables
(1)