-
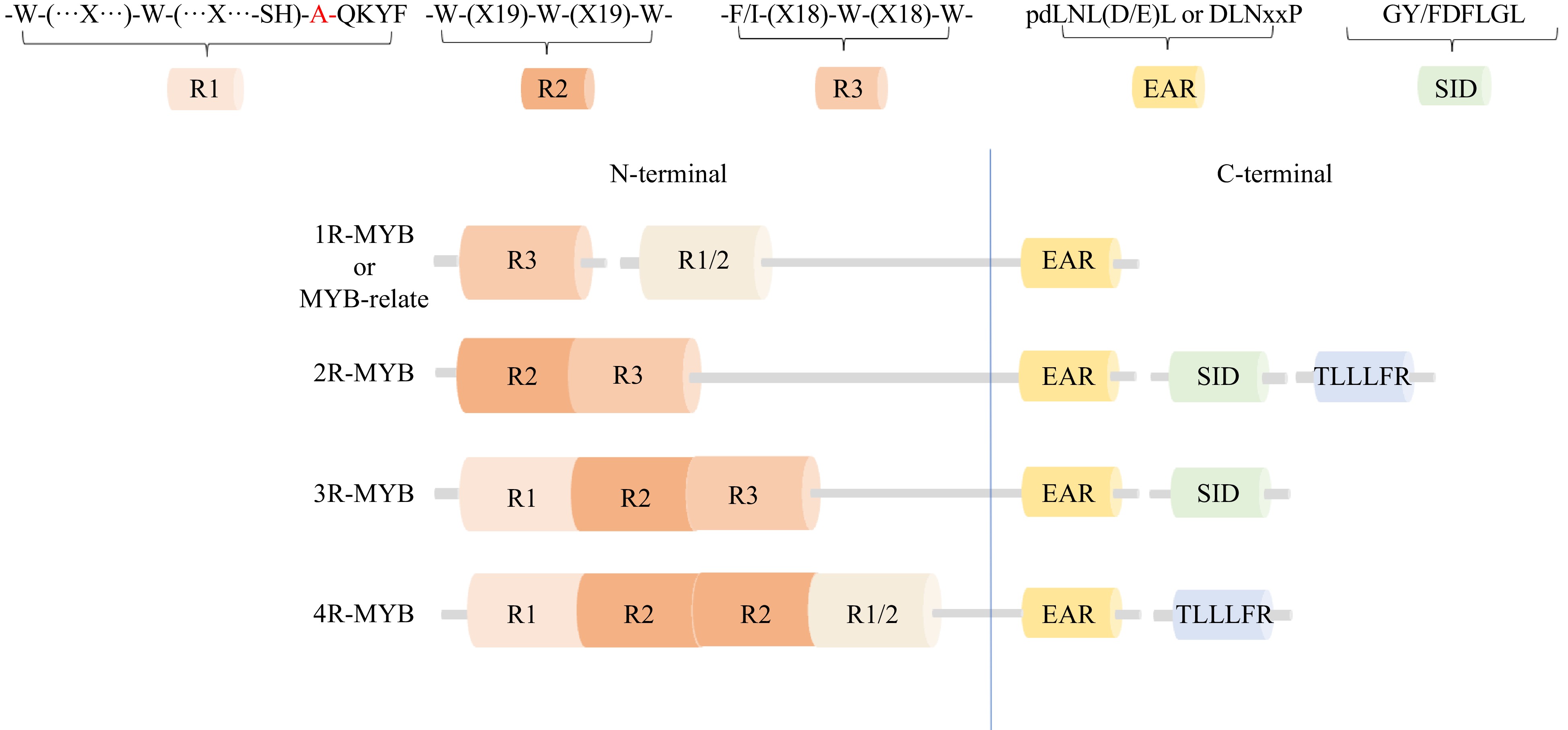
Figure 1.
Classification and structural characteristics of plant MYB transcription factors.
-
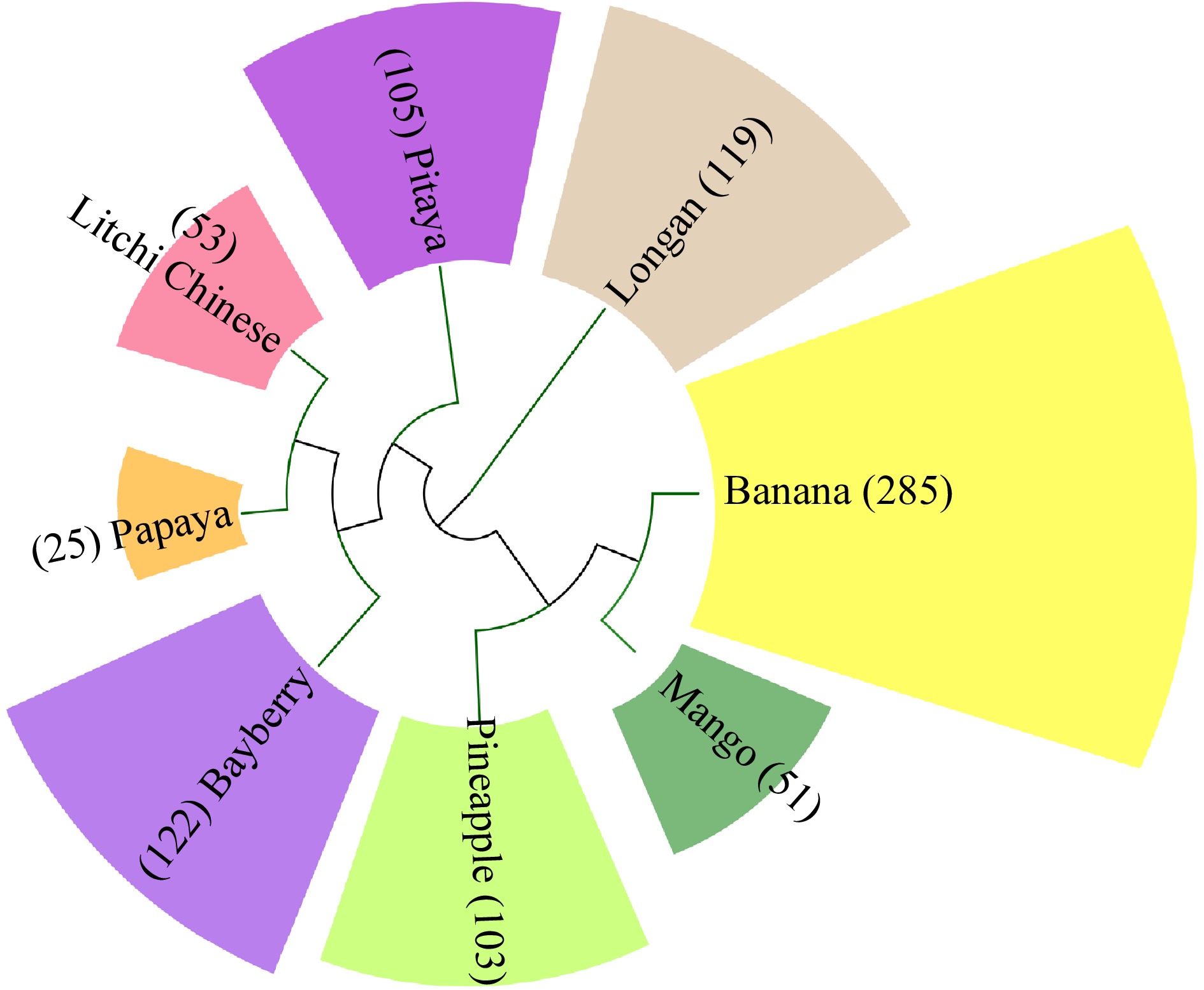
Figure 2.
Number of R2R3-MYB subfamily members in tropical fruit.
-
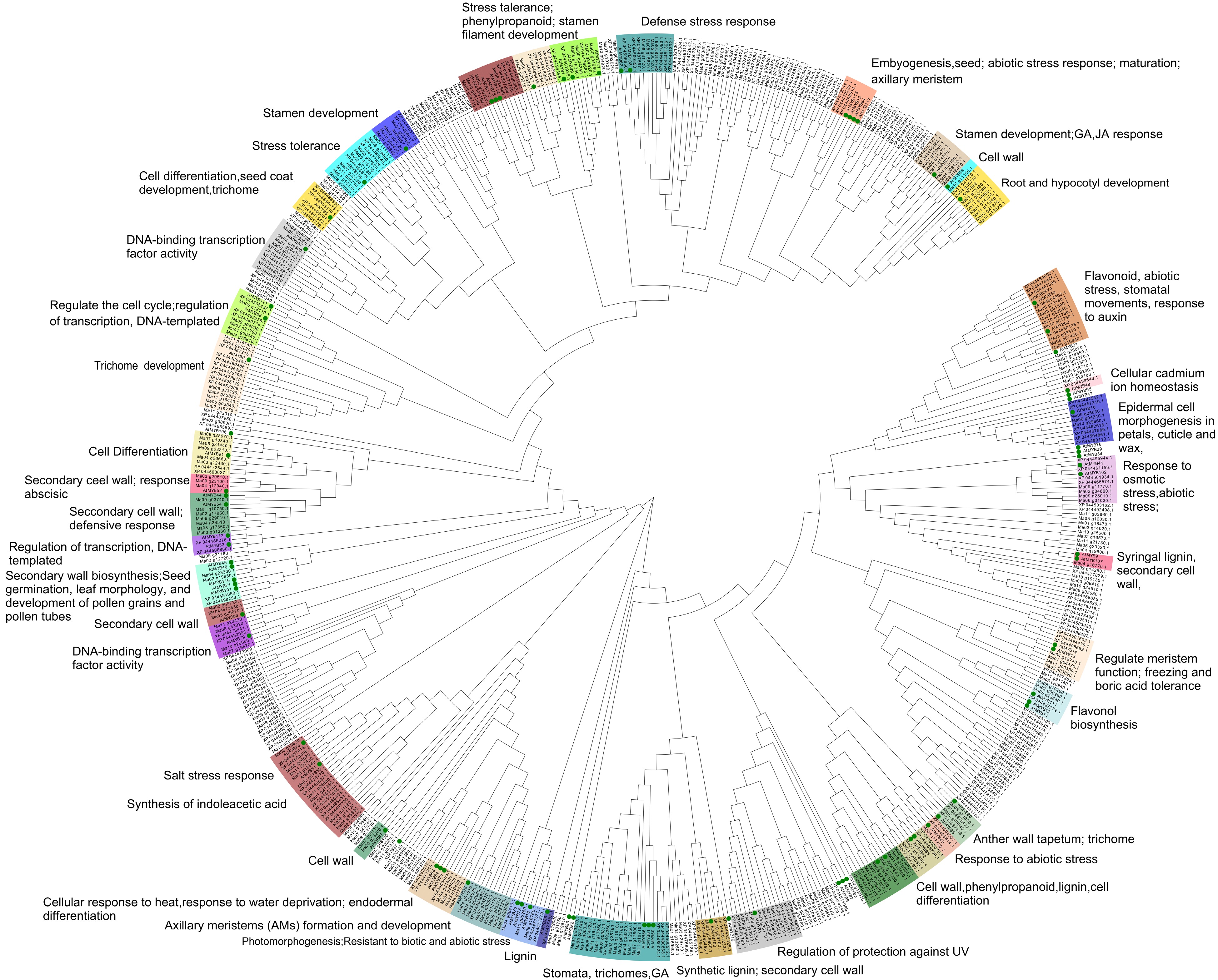
Figure 3.
Phylogenetic Neighbor-Joining (NJ) Tree.
-
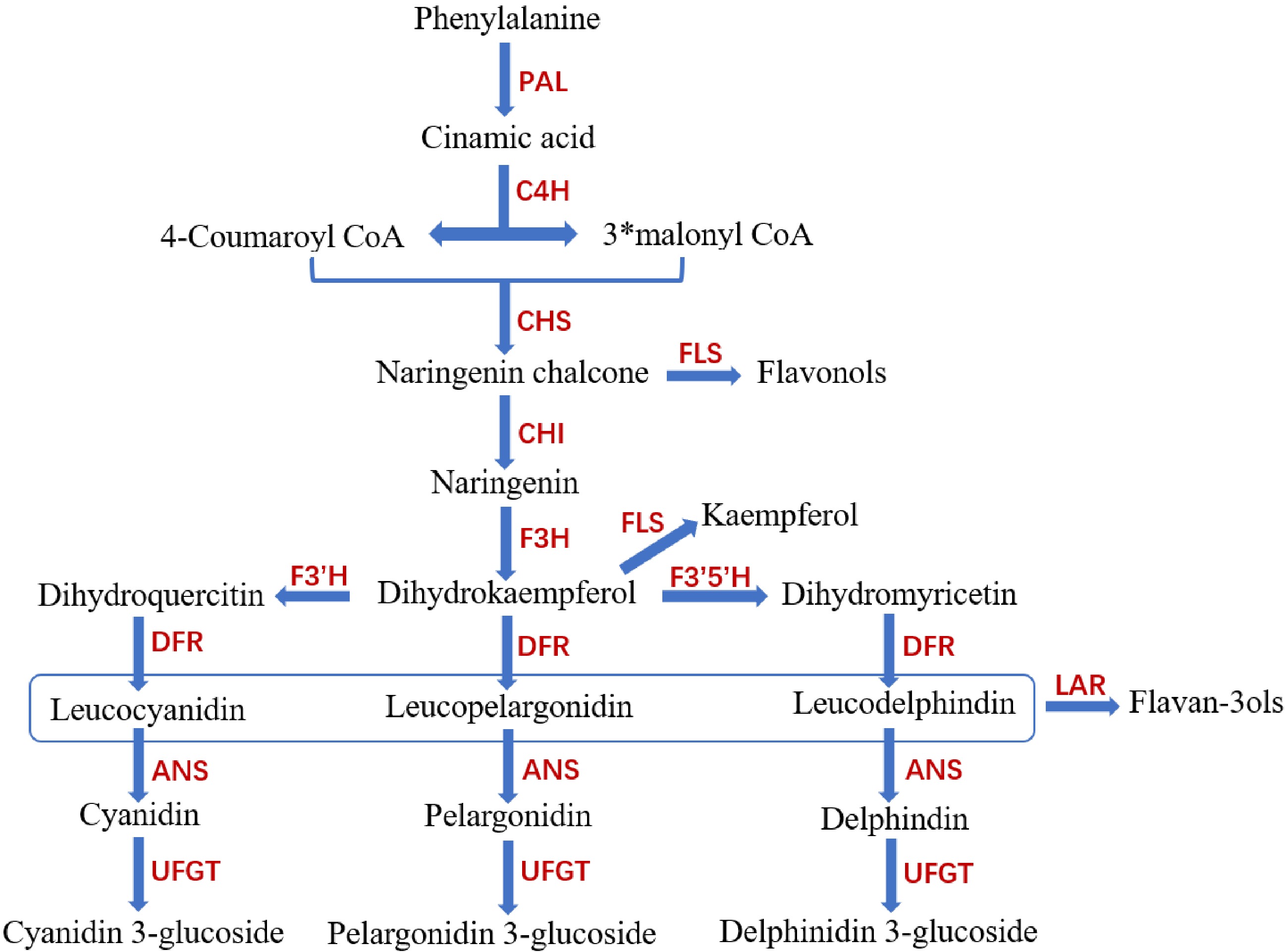
Figure 4.
Plant anthocyanin synthesis pathway. CHS (chalcone synthase), CHI (chalcone isomerase), F3H(flavanone 3-hydroxylase), F3′H (flavonoid 3′-hydroxylase Enzyme), DFR (dihydroflavonol 4-reductase), ANS (anthocyanin synthase) and UFGT (UDP glucose: flavonoid 3-O-glucosyltransferase), GST (Glutathione S-transferase), EAR (ERF-associated amphiphilic repression; Ethylene reaction factor related reaction inhibitory motifs).
-
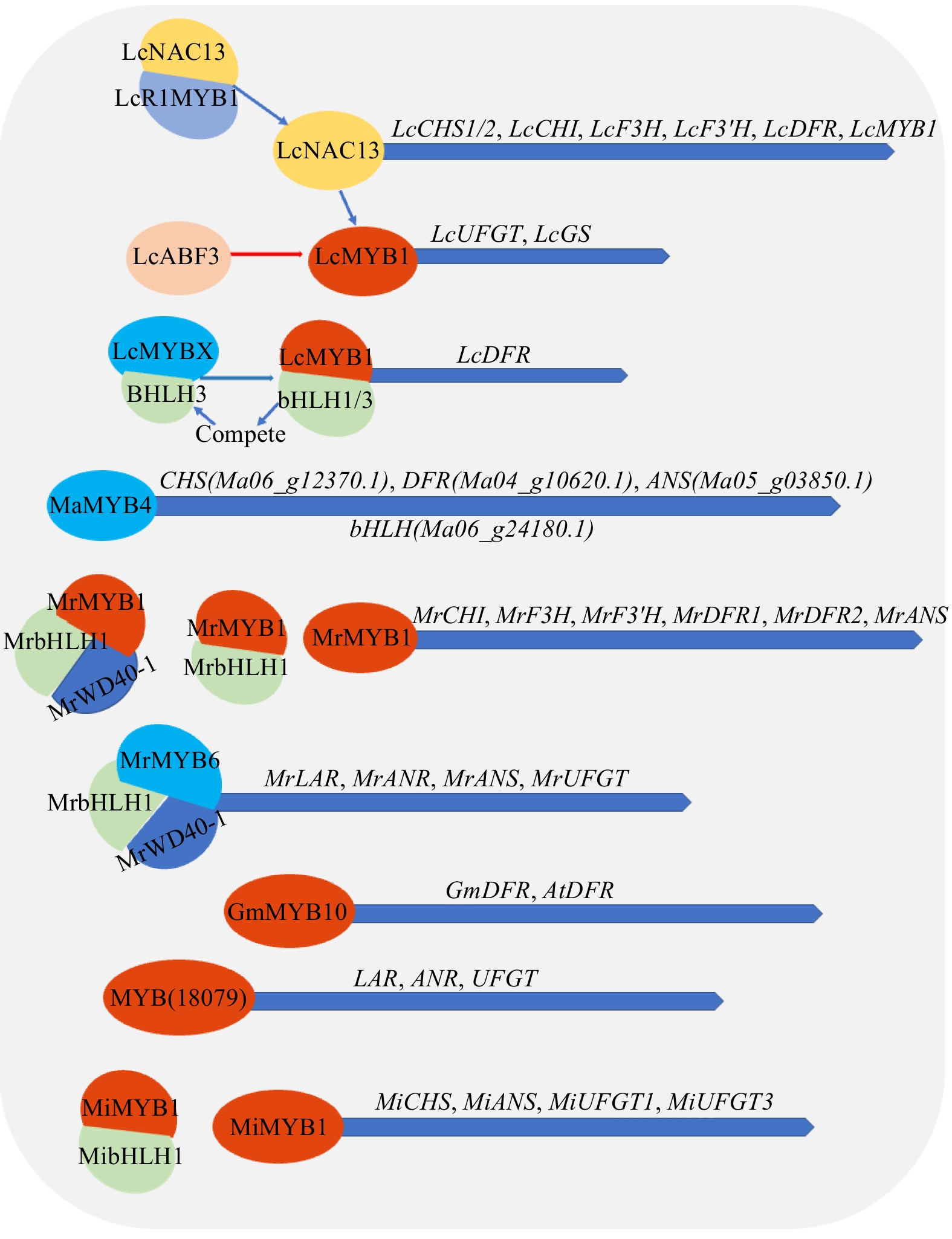
Figure 5.
Synthesis or decomposition of anthocyanins and flavonoids etc. by MYB transcription factor in tropical fruit. BHLH (basic helix-loop-helix), WD40 (WD40 repeat proteins), NAC (NAM, ATAF1, ATAF2 and CUC2). Note: blue represents inhibition, red represents activation and promotion.
-
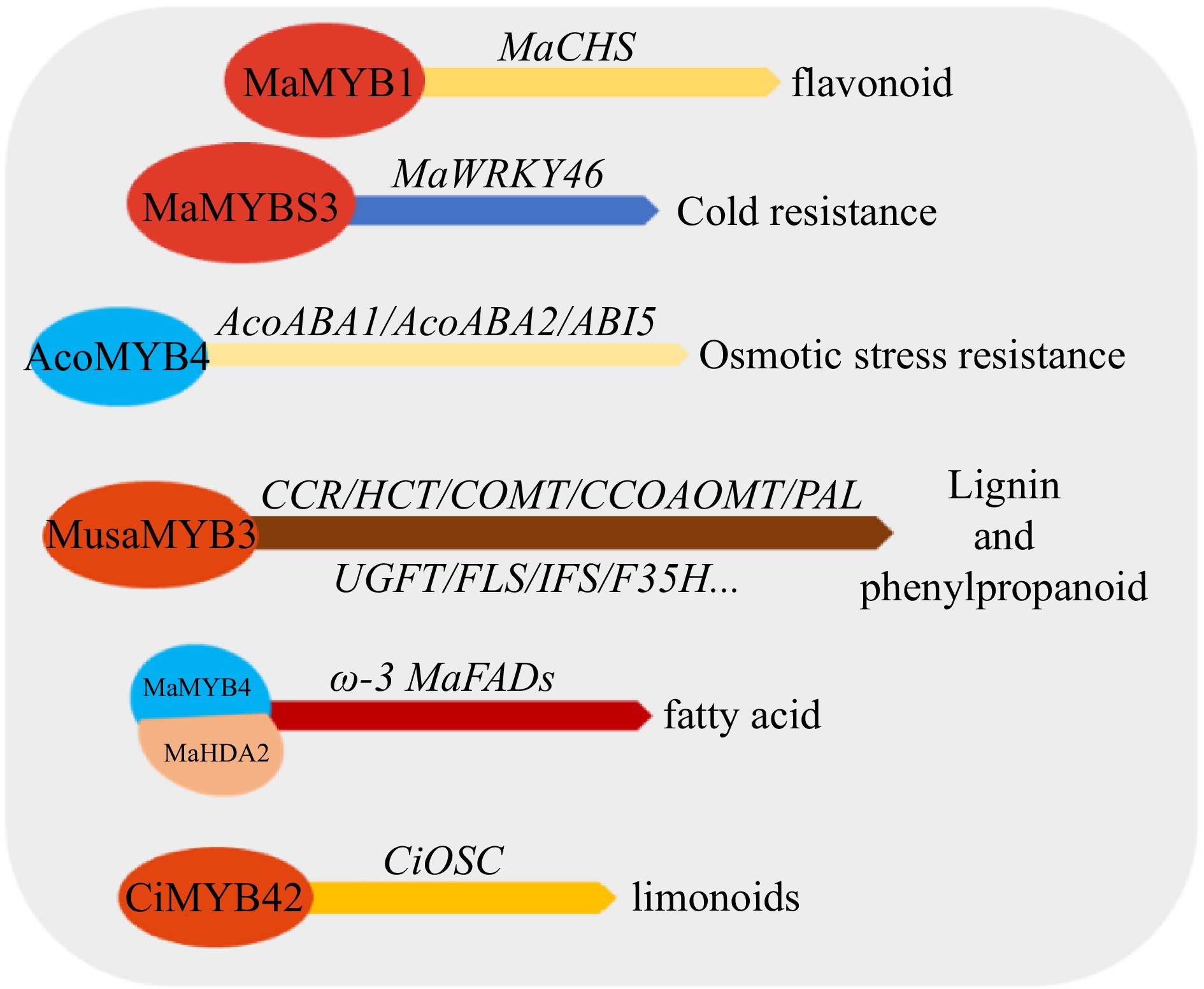
Figure 6.
Effects of MYB transcription factors on abiotic and biotic stress tolerance in tropical fruit. ABA1/ABA2 (genes responsible for ABA synthesis), ABI5 (Abscisic acid-insensitive 5, Key genes in response to ABA signals), CBF (C-repeat binding transcription factor/dehydrate responsive element binding factor, DREB), CHS (chalcone isomerase), FAD (fatty acid desaturase), CCR (Cinnamoyl CoA reductase), HCT (hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase), COMT (caffeic acid 3-O-methyltransferase), PAL (phenylalanine ammonia-lyase), CCOAOMT (caf-feoyl-CoA O-methyltransferase), UGFT (UDP-glucose: flavonoid-3-O-glucosyltransferase), FLS (flavonol synthase), IFS (isoflavone synthase), F35H (flavonoid 3,5-hydroxylases), HDA (histone deacetylase), OSC (Oxidosqualene cyclase).
-
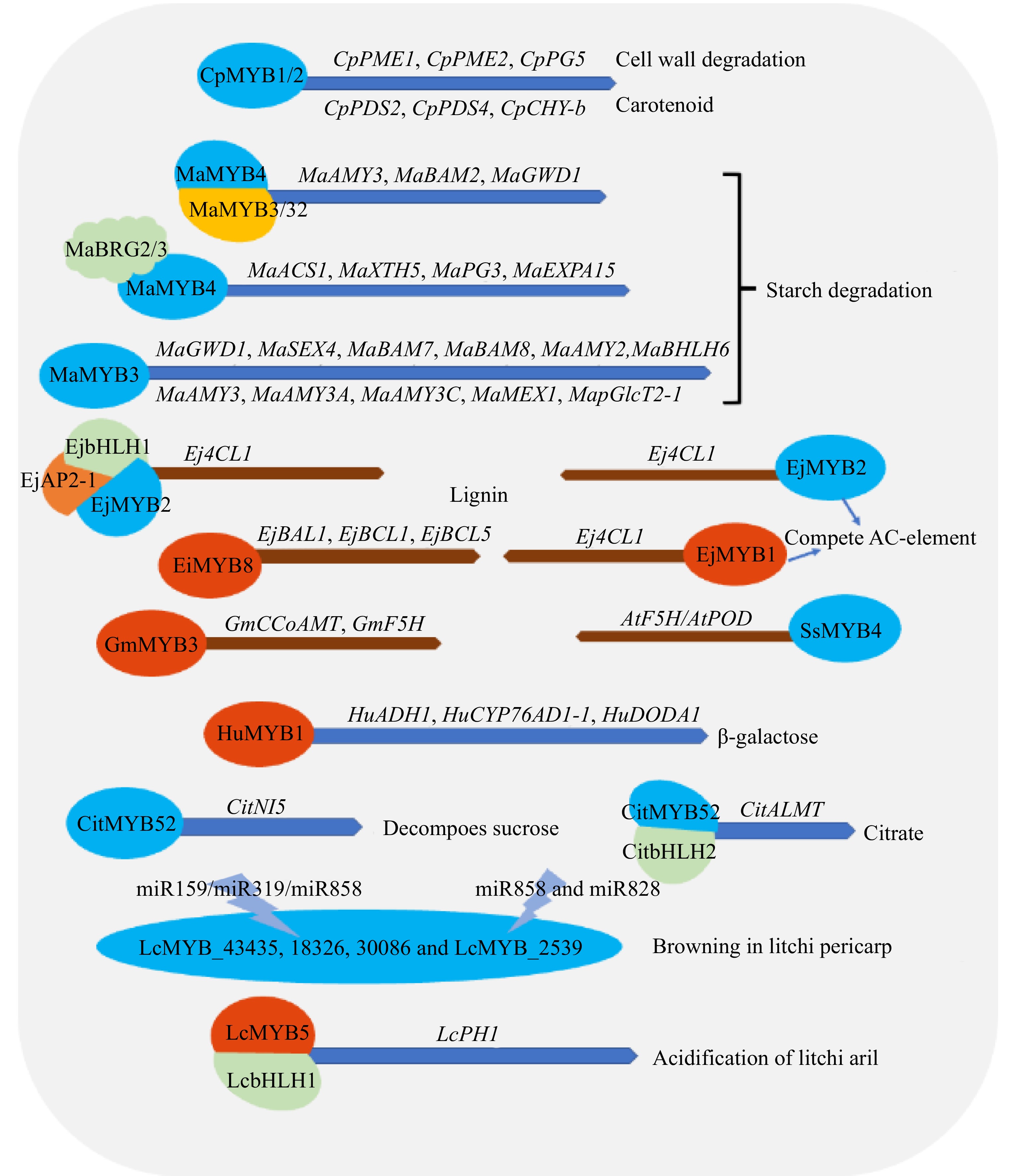
Figure 7.
Regulatory effects of MYB transcription factor on fruit ripening and fruit quality in tropical fruit crops. GWD (glucan-water dikinase), SEX4 (phosphoglucan phosphatases starch excess 4), BAM (Beta-amylase), AMY (α-amylases), MEX (maltose excess protein), pGlcT (plastidic glucose translocator), F5H (ferulate 5-hydroxylase), POD (Peroxidase), 4CL (4-coumarate;coenzyme A ligase), PAL (phenylalanine ammonia-lyase), PDS (Phytoene desaturase), CHY (Chymopapain), PME (pectinmethylesterase), PGs (polygalacturonases), NI (neutral/alkaline invertase), PGK (Phosphoglycerate kinase), Ca2+-ATPase (A multiregulated enzyme that is stimulated by calmodulin (CaM), KCI and lysophospholipids), ALMT (aluminum-activated malate transporter), ADH (Alcohol dehydrogenase), CYP76AD1 (cytochrome P 450 76AD1), DODA ((DOPA)-4,5-dioxygenase), AP2 (APETALA2), BRG2 (RING finger E3 ligases). MaACS1 (ACC synthase), MaXTH5 (cell wall modifying enzyme), MaPG3 (exopolymeric galacturonidase), MaEXPA15 (α-expansion protein) Note: blue represents inhibition, red represents activation and promotion.
-
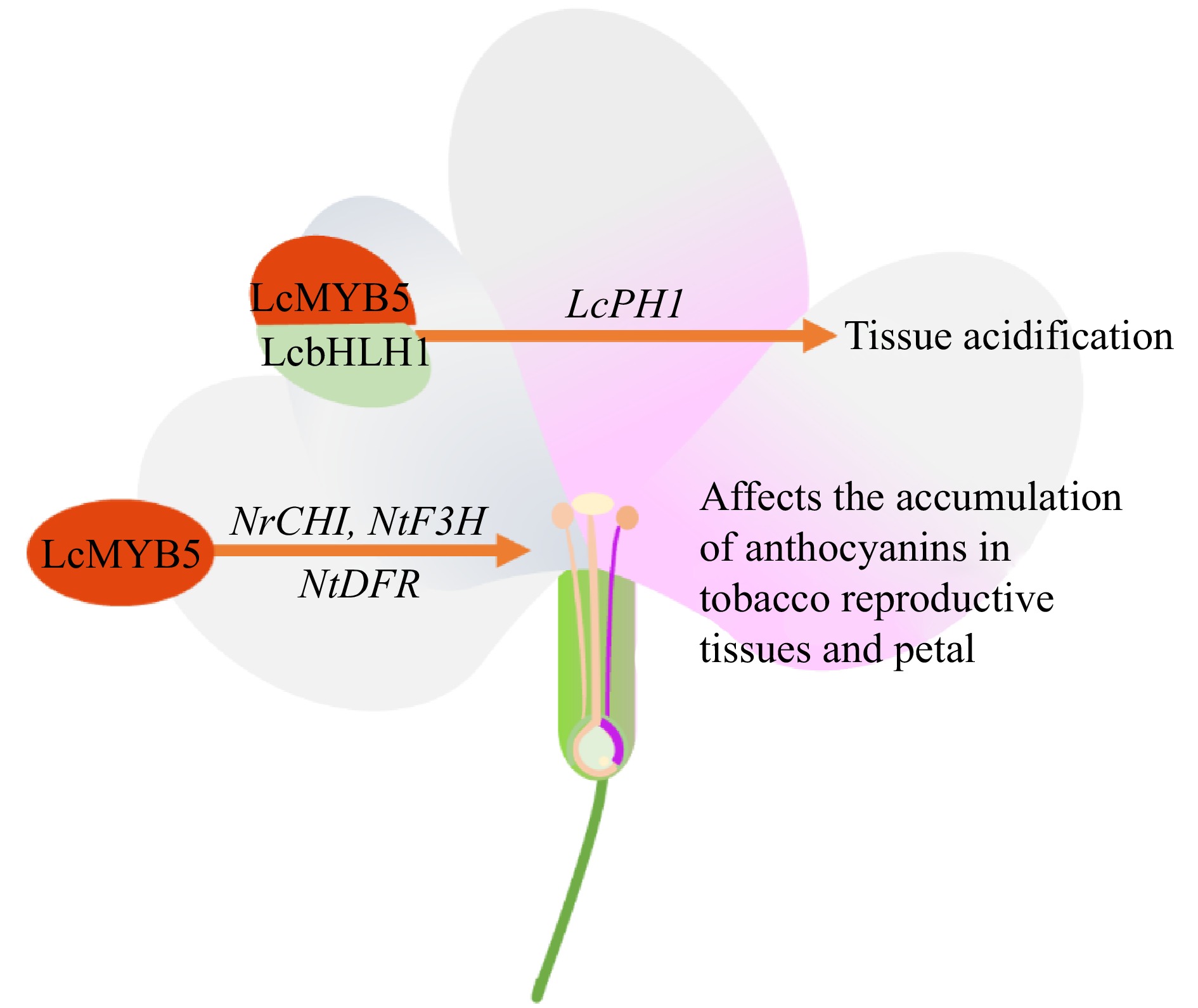
Figure 8.
Effects of MYB transcription factors on tropical fruit crops flowers. PH1 (P-ATPase), CHI (chalcone isomerase), F3H (flavanone 3-hydroxylase), DFR (dihydroflavonol 4-reductase).
-
Latin name Plant name Gene Gene founction Reference Litchi chinensis Litch LcMYB1 (KY302802.1) Regulates the accumulation of anthocyanins [68−72] LcR1MYB1 (QCF47524.1) Negative regulation of anthocyanin biosynthesis [73] LcMYB5 (Unigene 0002103) Anthocyanin biosynthesis, tissue acidification [127] LcMYBx (UCU01159.1) Negatively correlated with the synthesis of leaf anthocyanins [74] Mangifera indica Mango MinMYB10 Possible involvement in the regulation of anthocyanin accumulation in fruits [91] MinMYBPA Possible involvement in the regulation of anthocyanin accumulation in peels [92] MiMYB1 (NC_056567.1) Peel anthocyanin synthesis [93] Musa acuminata Banana MaMYB1 Response to drought, low temperature, salt stress [101] MaMYB14/63/110 Response process of salt stress [105] MaMYBR1 Resistance to fusarium wilt. [103] MaMYB3/4/32 Fruit ripening [119] MaMYB4 (Ma03_g21920) Inhibits banana anthocyanin biosynthesis and fruit ripeing; Negatively regulates the synthesis of banana Regulates the anthocyanins and synthesis of fatty acids [75,106,118] MaMYB3 Delay banana fruit ripening [116] MusaMYB31 Inhibits banana lignin and polyphenol biosynthesis [107] MaMYB16L Inhibited fruit ripening [115] MaMYB16S Promotes ripening of banana fruit [115] Musa spp. MpMYBS3 (KM379134) Participate in the regulation of cold signaling pathways. [108] Musa paradisiaca MYB108 (Ma09_g15940) Regulates the growth and development of banana leaves and roots under long-term magnesium deficiency conditions [104] Ananas comosus Pineapple AcoMYB1 (XM_020230319) Regulates early development and late ripening in pineapples in response to drought and low temperature stress [109] AcoMYB4 Inhibit ABA synthesis [110] Hylocereus polyrhizus Pitaya HuMYB1814 Synthesis of Anthocyanins [83] Hylocereus undatus Unigene0036816 May be to relevant the color difference of dragon fruit pulp [84] Unigene0032395 May be to relevany the color difference of dragon fruit pulp [84] Unigene0034590 May be to relevant the color difference of dragon fruit pulp [84] Hylocereus polyrhizus HuMYB1 Inhibits the expression of β-galactose-related genes. [121] Garcinia mangostana Mangosteen GmMYB10 (ACM62751.1) Regulation of anthocyanin biosynthesis before and after fruit harvest [85] GmMYB30 (AID69234.1) Cell wall thickening, lignin
biosynthesis and stress response.
[128]Myrica rubra Bayberry MrMYB1 (GQ340767) Expression is induced by light signals, which then affects anthocyanin accumulation [76−80] MrMYB6 (c24596_g1) May negatively regulate the anthocyanins and PA (proanthocyanidin)accumulation [81] Macadamia
integrifoliaMacadamia nuts MiMYB1 (QIJ70646.1) Involved in stress response [111] MiMYB2 (QIJ70647.1) Related to growth [133] Carica papaya Papaya CPMYB1 Fruit softening, carotenoid accumulation, fruit ripening [120] CPMYB-2 Fruit softening, carotenoid accumulation, fruit ripening [120] Theobroma cacao Cocoa TcMYB113 Regulation of flavonoid synthesis genes to determine red pod color in cocoa beans [86] Tc-MYBPA Involved in the synthesis of anthocyanins and procyanidins in cocoa [88] MYB(gene18079) Involved in the regulation of cocoa pod color [87] Citrus unshiu Citrus CitMYB52 Transcriptional regulation involved in sucrose metabolism [11] Citrus reticulata CitMYB52 Negatively regulate citrate accumulation, Inhibits the breakdown of sucrose [121] Fortunella japonica CitMYB20 (MN689609) Associated with citrus canker resistance [112] C. reticulata/C. sinensis CiMYB42 (Ciclev10021695m) Involved in the biosynthesis of citrus limonoids [113] Dimocarpus Longan Longan MYB-related-R1 May be related to chromosome telomere structure and anthocyanin synthesis [89] MYB14 May be to relevant the flowering process of longan [134] Syzygium samarangense Wax apple SsMYB1/4/20/54/77 (Transcriptome database:PRJNA284092) Promote lignin synthesis [63] SsMYB108/114/308 Promote lignin synthesis [63] SsMYB3/5/6/10/33/39
(comp40847_c1/
comp186052_c0/
comp17591_c0/
comp_45853_c0/
comp_44103_c0Promote lignin synthesis [63] SsMYB44/48/86/306/330/C1 Related to postharvest lignin synthesis [63] SsMYB48 Inhibit lignin synthesis [63] Eriobotrya japonica Loquat EjMYB1 (KF767453) Fruit chilling lignification [123,124] EjMYB2 (KF767454) Lignification Negative Regulator [43,123,124] EjMYB8 Promote lignin synthesis [125] EjMYB9 Promote lignin synthesis [125] Table 1.
MYB genes and their main functions in tropical fruit.
Figures
(8)
Tables
(1)