-

Figure 1.
Transverse cross-sections of apple slices showing range in (a) flesh colouration, and (b) flesh browning disorder for Type 1 red-fleshed apple. The weighted cortical intensity (WCI) scores (0−9 scale) and the proportion of the cortex area showing symptoms of flesh browning disorder are also displayed.
-
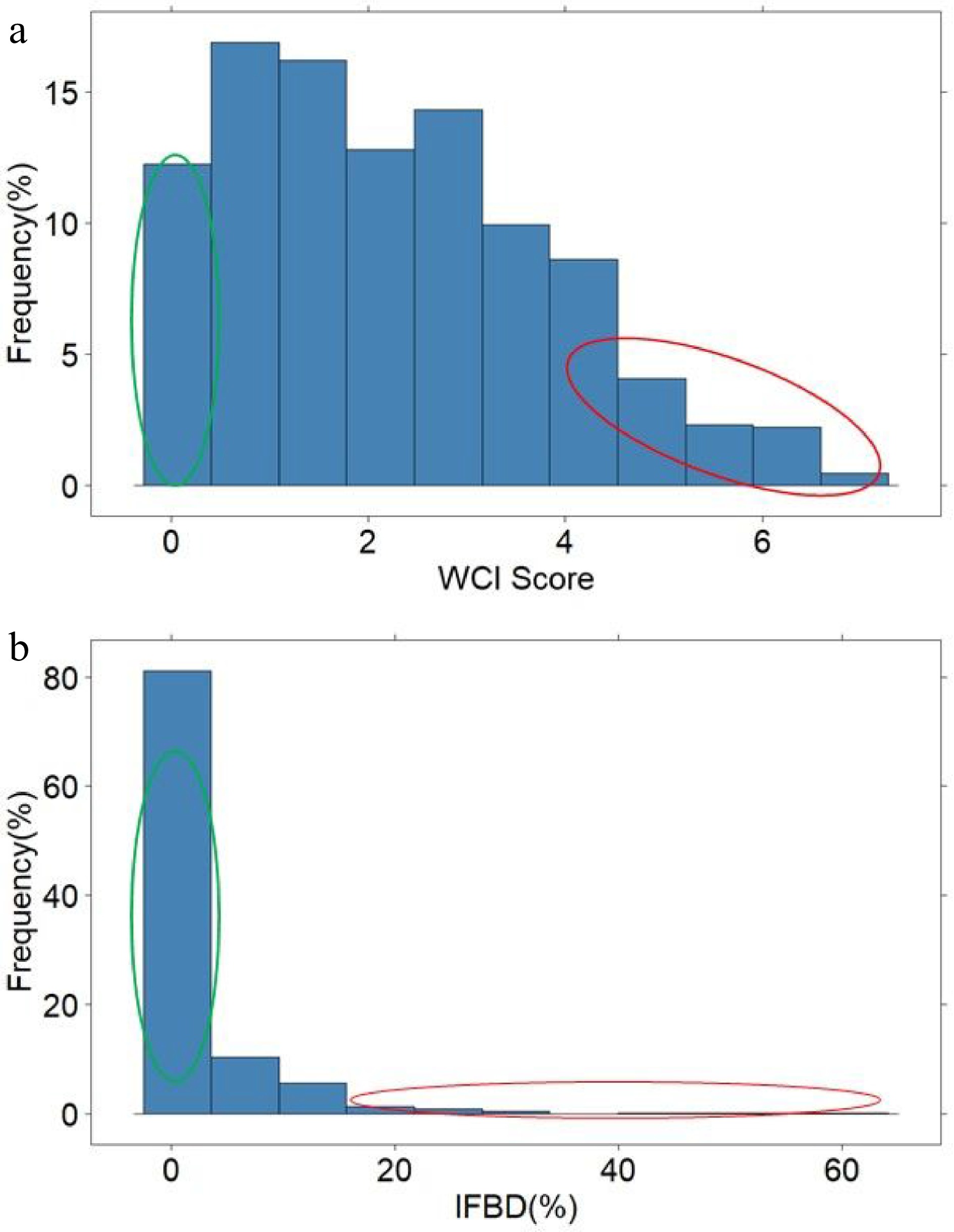
Figure 2.
The distribution of weighted cortex intensity (WCI) scores (a) and the internal flesh browning disorder IFBD%; (b) in the population of ~900 apple seedlings. The green and red circles highlight the individuals used to form the 'low' and 'high' pools of samples.
-
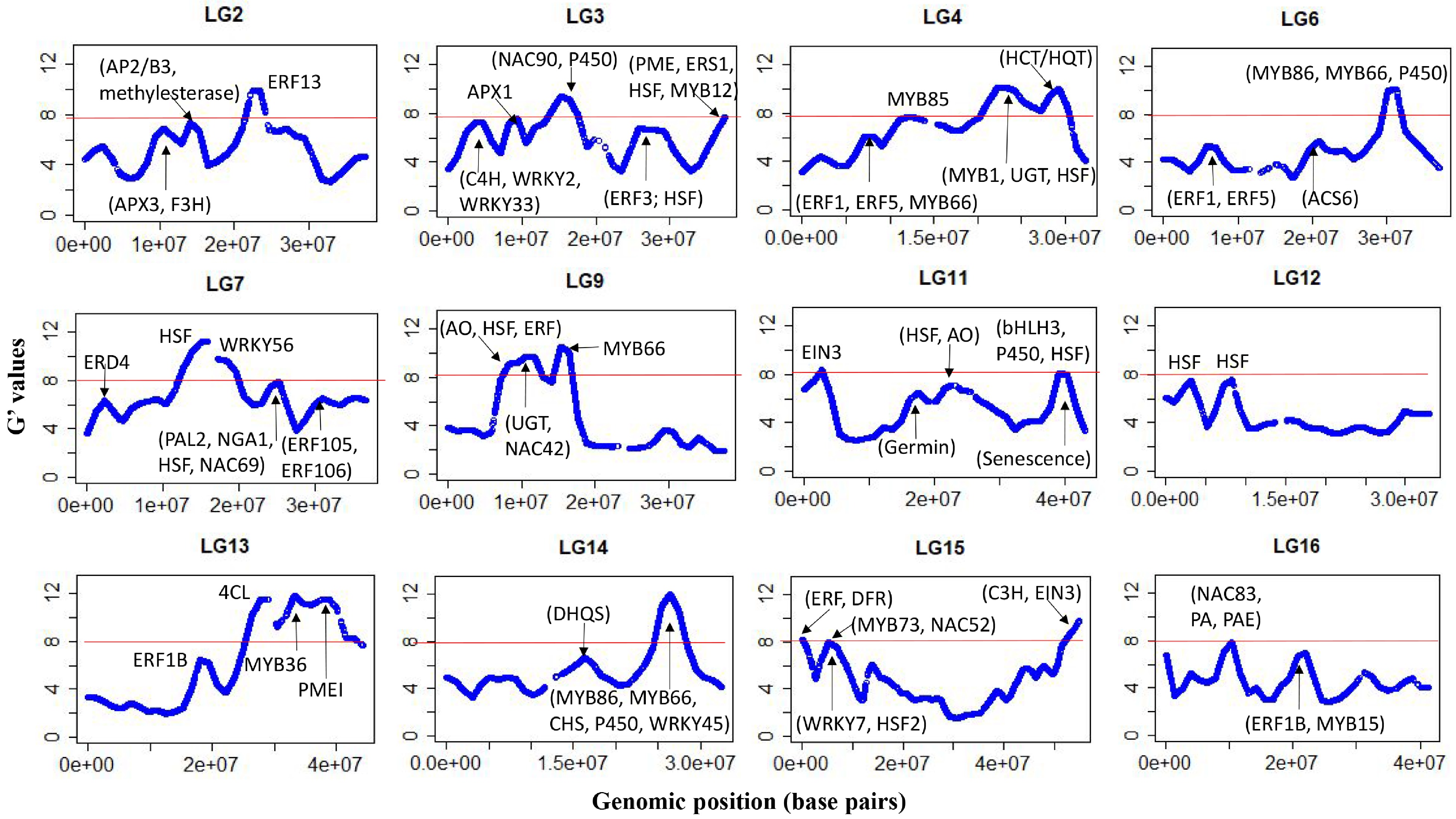
Figure 3.
G'-statistics across the linkage groups (LG) showing significant association with the flesh browning disorder (FBD) in apple The horizontal red lines indicate the significance threshold. The putative candidate genes (refer to Table 1) underpinning various G' peaks are also shown.
-
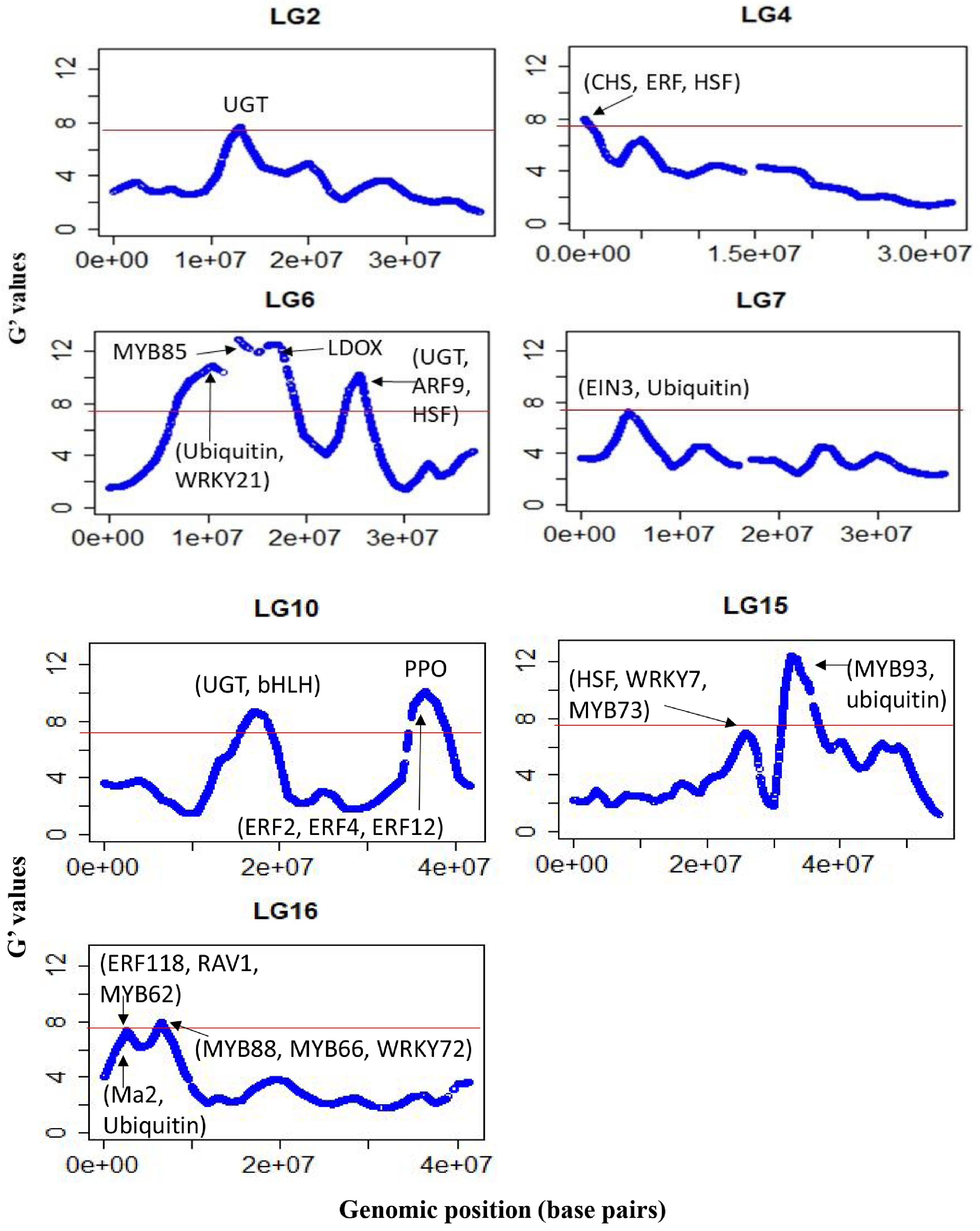
Figure 4.
G’-statistics across the linkage groups (LG) showing significant association with the weighted cortex intensity (WCI) in apple. The horizontal red lines indicate the significance threshold. The putative candidate genes (refer to Table 2) underpinning various G' peaks are also shown.
-
Chr Genomic region (Mb) Putative genes functions 2 21.9–23.5 Ethylene-responsive element binding factor 13 (MdERF13: MD02G1213600); 3 3.1–4.9 cinnamate 4-hydroxylase (C4H) enzyme (MD03G1051100, MD03G1050900 and MD03G1051000); MdWRKY2: MD03G1044400; MdWRKY33 (MD03G1057400) 3 8.2–9.6 ascorbate peroxidase 1 (MdAPX1: MD03G1108200, MD03G1108300) 3 14.7–16.6 senescence-related MdNAC90 (MD03G1148500) 3 36.5–37.5 Ethylene response sensor 1 (MdERS1: MD03G1292200); flavonoid biosynthesis protein MdMYB12 (MD03G1297100); Heat shock protein DnaJ (MD03G1296600, MD03G1297000); pectin methylesterase (MdPME) inhibitor protein (MD03G1290800, MD03G1290900, MD03G1291000). 4 11.0–13.0 phenylalanine and lignin biosynthesis protein MdMYB85 (MD04G1080600) 4 22.1–24.4 MYB domain protein 1 (MD04G1142200); HSP20-like protein (MD04G1140600); UDP-glucosyltransferase (UGT) proteins UGT85A7 (MD04G1140700, MD04G1140900); UGT85A3 (MD04G1140800); UGT (MD04G1141000, MD04G1141300); UGT85A2 (MD04G1141400); UGT85A4 (MD04G1141500); DNAJ heat shock protein (MD04G1153800, MD04G1153900, MD04G1154100) 4 27.6–29.6 HCT/HQT regulatory genes MD04G1188000 and MD04G1188400 6 29.8–31.7 Volz et al. (2013) QTL for IFBD; anthocyanin regulatory proteins MdMYB86 (MD06G1167200); triterpene biosynthesis transcription factor MdMYB66 (MD06G1174200); Cytochrome P450 (MD06G1162600; MD06G1162700, MD06G1162800, MD06G1163100, MD06G1163300, MD06G1163400, MD06G1163500, MD06G1163600, MD06G1163800, MD06G1164000, MD06G1164100, MD06G1164300, MD06G1164400, MD06G1164500, MD06G1164700) 7 13.9–15.9 heat shock protein 70B (MD07G1116300) 7 17.4–19.0 Drought-stress WRKY DNA-binding proteins (MdWRKY56: MD07G1131000, MD07G1131400) 7 23.6–25.2 MdPAL2 (MD07G1172700); drought-stress gene NGA1 (MD07G1162400); DNAJ heat shock family protein (MD07G1162300, MD07G1162200), stress-response protein (MdNAC69: MD07G1163700, MD07G1164000) 9 7.9–11.8 MD09G1110500 involved in ascorbate oxidase (AO); MdUGT proteins (MD09G1141200, MD09G1141300, MD09G1141500, MD09G1141600, MD09G1141700, MD09G1141800, MD09G1142000, MD09G1142500, MD09G1142600, MD09G1142800, MD09G1142900, MD09G1143000, MD09G1143200, MD09G1143400) involved in flavonoids biosynthesis; heat shock proteins 89.1 (MD09G1122200) and HSP70 (MD09G1137300); Ethylene-forming enzyme MD09G1114800; Anthocyanin regulatory protein MdNAC42 (MD09G1147500, MD09G1147600) 9 14.9–16.5 triterpene biosynthesis transcription factor protein MdMYB66 (MD09G1183800); 11 1.7–3.0 ethylene response factor proteins (MdEIN-like 3: MD11G1022400) 11 38.6–40.6 Senescence-related gene 1 (MD11G1271400, MD11G1272300, MD11G1272000, MD11G1272100, MD11G1272300, MD11G1272400, and MD11G1272500); Chalcone-flavanone isomerase (CHI) protein (MD11G1273600) and MdbHLH3 (MD11G1286900, MDP0000225680); cytochrome P450 enzyme (MD11G1274000, MD11G1274100, MD11G1274200, MD11G1274300, MD11G1274500, and MD11G1274600); heat shock transcription factor A6B (MdHSFA6B: MD11G1278900) – involved in ABA-mediated heat response and flavonoid biosynthesis. 12 2.2–3.5 heat shock protein 70-1 ((MD12G1025600, MD12G1025700 and MD12G1026300) and heat shock protein 70 (MD12G1025800 and MD12G1025900 and MD12G1026000); ethylene (MD12G1032000) and auxin-responsive (MD12G1027600) proteins. 12 7.2–8.4 DNAJ heat shock domain-containing protein (MD12G1065200 and MD12G1067400) 13 27.5–30.5 MD13G1257800 involved in polyphenol 4-coumarate:CoA ligase (4CL) 13 37.5–39.5 pectin methyl esterase inhibitor superfamily protein MdPMEI (MD13G1278600) 14 25.4–27.5 Drought-stress response gene MdWRKY45 (MD14G1154500); chalcone synthase (CHS) family proteins (MD14G1160800 and MD14G1160900); triterpene biosynthesis transcription factor MdMYB66 (MD14G1180700, MD14G1181000, MD14G1180900); anthocyanin biosynthesis protein (MdMYB86: MD14G1172900); cytochrome P450 proteins (MD14G1169000, MD14G1169200, MD14G1169600, MD14G1169700) 15 0–1.5 Ethylene synthesis proteins (MD15G1020100, MD15G1020300 and MD15G1020500); dihydroflavonol reductase (DFR) gene (MD15G1024100) 15 4.6–6.8 MdMYB73 (MD15G1076600, MD15G1088000) modulates malate transportation/accumulation via interaction with MdMYB1/10; MdNAC52: MD15G1079400) regulates anthocyanin/PA; heat shock transcription factor B4 (MD15G1080700); stress-response protein MdWRKY7 (MD15G1078200) 15 53.5–54.9 MdC3H (MD15G1436500) involved in chlorogenic acid biosynthesis; MdEIN3 (MD15G1441000) involved in regulating ethylene synthesis and anthocyanin accumulation 16 8.9–10.9 SAUR-like auxin-responsive protein (MD16G1124300) and MdNAC83: (MD16G1125800) associated with fruit ripening; MD16G1140800 regulates proanthocyanidin; MdPAE genes (MD16G1132100, MD16G1140500) regulates ethylene production. Table 1.
A list of the genomic regions associated with internal flesh browning disorder (FBD) in apples. Putative candidate genes residing within these regions are also listed using GDDH13v1.1 reference genome assembly.
-
Chr Genomic region (Mb) Putative genes functions 2 11.2–13.2 UDP-glucosyltransferase (UGT) proteins (MD02G1153000, MD02G1153100, MD02G1153200, MD02G1153300; MD02G1153400; MD02G1153500; MD02G1153700; MD02G1153800; MD02G1153900); 4 0–1.2 Chalcone synthase (CHS) genes (MD04G1003000; MD04G1003300 and MD04G1003400); DNAJ heat shock protein (MD04G1003500); MdERF (MD04G1009000) involved in regulating PA biosynthesis. 6 9.5–11.0 Ubiquitin protein (MD06G1061100); stress-response WRKY protein MdWRKY21 (MD06G1062800); 6 12.1–13.6 pectin methylesterase inhibitor superfamily protein (MdPME: MD06G1064700); phenylalanine and lignin biosynthesis gene (MdMYB85) 6 16.0–17.6 MD06G1071600 (MDP0000360447) involved in leucoanthocyanidin dioxygenase (LDOX) synthesis 6 24.0–26.0 UDP-glycosyltransferase proteins (MD06G1103300, MD06G1103400, MD06G1103500 and MD06G1103600); auxin response factor 9 (MdARF9: MD06G1111100) and heat shock protein 70 (Hsp 70; MD06G1113000). 7 4.1–5.6 Ethylene insensitive 3 protein (MdEIN3: MD07G1053500 and MD07G1053800) involved in proanthocyanidins (PA) biosynthesis; ubiquitin-specific protease (MD07G1051000, MD07G1051100, MD07G1051200, MD07G1051500, MD07G1051300, MD07G1051700 and MD07G1051800). 10 15.9–18.3 bHLH proteins (MD10G1098900, MD10G1104300, and MD10G1104600); UGT protein (MD10G1101200); UGT 74D1 (MD10G1110800), UGT 74F1 (MD10G1111100), UGT 74F2 (MD10G1111000). 10 35.6–38.6 Stress-response WRKY proteins (MdWRKY28: MD10G1266400; MdWRKY65: MD10G1275800). ethylene responsive factors (MdERF2: MD10G1286300; MdERF4: MD10G1290400, and MdERF12: MD10G1290900) and a NAC domain protein (MdNAC73: MD10G1288300); polyphenol oxidase (PPO) genes ((MD10G1298200; MD10G1298300; MD10G1298400; MD10G1298500; MD10G1298700; MD10G1299100; MD10G1299300; MD10G1299400). 15 24.8–26.8 heat shock factor 4 (MD15G1283700), drought-stress response WRKY protein 7 (MdWRKY7: MD15G1287300), MdMYB73 (MD15G1288600) involved in ubiquitination and malate synthesis 15 31.7–34.2 MYB domain protein 93 (MdMYB93: MD15G1323500) regulates flavonoids and suberin accumulation (Legay et al. 2016); ubiquitin -specific protease 3 (MD15G1318500), 16 1.5–3.4 MYB domain proteins (MD16G1029400) regulates anthocyanin; senescence-associated gene 12 (MD16G1031600); ethylene response factor proteins (MdERF118: MD16G1043500, and MD16G1047700 (MdRAV1); MdMYB62 ( MD16G1040800) flavonol regulation; malate transporter MdMa2 (MD16G1045000: MDP0000244249), Ubiquitin-like superfamily protein (MD16G1036000), 16 5.4–7.4 MdMYB88 (MD16G1076100) regulates phenylpropanoid synthesis and ABA-mediated anthocyanin biosynthesis; MdMYB66 (MD16G1093200) regulates triterpene biosynthesis, MdWRKY72 (MD16G1077700) mediates ultraviolet B-induced anthocyanin synthesis. Table 2.
A list of the genomic regions significantly associated with the weighted cortex intensity (WCI) in apples. Putative candidate genes residing within these regions are also listed using GDDH13v1.1 reference genome assembly.
-
Trait Chr (genomic region: Mb) Gene name Predicted gene ID Putative function IFBD Chr6 (29.8–31.7 Mb) MdMYB66 MD06G1174200 Suberin/triterpene deposition IFBD Chr14 (25.4–27.5 Mb) MdMYB66 MD14G1180700, MD14G1181000, MD14G1180900 Suberin/triterpene deposition IFBD Chr4 (7.0–8.1 Mb) MdMYB66 MD04G1060200 Suberin/triterpene deposition IFBD Chr9 (14.9–16.5 Mb) MdMYB66 MD09G1183800 Suberin/triterpene deposition IFBD Chr6 (29.8–31.7 Mb) MdMYB86 MD06G1167200 Anthocyanin regulation IFBD Chr14 (25.4–27.5 Mb) MdMYB86 MD14G1172900 Anthocyanin regulation IFBD Chr6 (29.8–31.7 Mb) MdMYB98 MD06G1172900 Drought stress response IFBD Chr14 (25.4–27.5 Mb) MdMYB98 MD14G1179000 Drought stress response IFBD Chr6 (29.8–31.7 Mb) Cytochrome P450 MD06G1162600, MD06G1162700, MD06G1162800, MD06G1163100, MD06G1163300, MD06G1163400, MD06G1163500, MD06G1163600, MD06G1163800, MD06G1164000, MD06G1164100, MD06G1164300, MD06G1164400, MD06G1164500, MD06G1164700 Flavonoid and triterpenic metabolism IFBD Chr14 (25.4–27.5 Mb) Cytochrome P450 MD14G1169000, MD14G1169200, MD14G1169600, MD14G1169700 Flavonoid and triterpenic metabolism IFBD Chr3 (14.7–16.6 Mb) MdNAC90 MD03G1148500 Senescence-related IFBD Chr11 (16.5–18.5 Mb) MdNAC90 MD11G11679000 Senescence-related IFBD Chr14 (25.4–27.5 Mb) MdNAC83 MD14G1150900 Senescence/ripening-related IFBD Chr16 (8.9–10.9 Mb) MdNAC83 MD16G1125800 Senescence/ripening-related IFBD Chr17 (0–0. 7 Mb) MdNAC83 MD17G1010300 Senescence/ripening-related IFBD Chr3 (14.7–16.6 Mb) Germin-like protein 10 MD03G1148000 Polyphenol oxidase IFBD Chr11 (16.5–18.5 Mb) Germin-like protein 10 MD11G1167000, MD11G1167100, MD11G1167400, MD11G1169200 Polyphenol oxidase IFBD Chr1 (26.9–28.5 Mb) MdWRKY55 MD01G1168500 Drought stress response IFBD Chr7 (30.3–31.9 Mb) MdWRKY55 MD07G1234600 Drought stress response IFBD Chr1 (26.9–28.5 Mb) MdWRKY70 MD01G1168600 Drought stress response IFBD Chr7 (30.3–31.9 Mb) MdWRKY70 MD07G1234700 Drought stress response IFBD Chr4 (7.0–8.1 Mb) MdERF1 MD04G1058000 Ethylene responsive factor IFBD Chr6 (5.5–7.3 Mb) MdERF1 MD06G1051800 Ethylene responsive factor IFBD Chr4 (7.0–8.1 Mb) MdERF5 MD04G1058200 Ethylene responsive factor IFBD Chr6 (5.5–7.3 Mb) MdERF5 MD06G1051900 Ethylene responsive factor IFBD Chr13 (18.0–20.0 Mb) MdERF1B MD13G1213100 Ethylene response factor 1 IFBD Chr16 (20.4–22.4 Mb) MdERF1B MD16G1216900 Ethylene response factor 1 WCI Chr13 (2.6–4.4 Mb) MdRAV1 MD13G1046100 Ethylene responsive factor WCI Chr16 (1.5–3.4 Mb) MdRAV1 MD16G1047700 Ethylene responsive factor WCI Chr13 (2.6–4.4 Mb) MdMYB62 MD13G1039900 Flavonol biosynthesis WCI Chr16 (1.5–3.4 Mb) MdMYB62 MD16G1040800 Flavonol biosynthesis IFBD Chr9 (7.9–11.8 Mb) MdNAC42 MD09G1147500, MD09G1147600 Anthocyanin accumulation WCI Chr17 (11.4–12.4 Mb) MdNAC42 MD17G1134400 Anthocyanin accumulation IFBD Chr9 (7.9–11.8 Mb) HSP 70 MD09G1137300 Heat stress response WCI Chr17 (11.4–12.4 Mb) HSP 70 MD17G1127600 Heat stress response IFBD Chr4 (11.0–13.0 Mb) MdMYB85 MD04G1080600 Phenylalanine and lignin biosynthesis WCI Chr6 (12.1–13.6 Mb) MdMYB85 MD06G1064300 Phenylalanine and lignin biosynthesis IFBD Chr15 (13.2–14.2 Mb) MdEBF1 MD15G1171800 Ethylene inhibition WCI Chr8 (15.2–17.2 Mb) MdEBF1 MD08G1150200 Ethylene inhibition IFBD Chr15 (53.5–54.9 Mb) MdEIN3 MD15G1441000 Ethylene insensitive 3 protein WCI Chr8 (30.3–31.6 Mb) MdEIN3 MD08G1245800 Ethylene insensitive 3 protein IFBD Chr11 (1.7–3.0 Mb) MdEIN3 MD11G1022400 Ethylene insensitive 3 protein WCI Chr7 (4.1–5.6 Mb) MdEIN3 MD07G1053500, MD07G1053800 Ethylene insensitive 3 protein IFBD Chr15 (4.6–6.8 Mb) MdMYB73 MD15G1076600, MD15G1088000 Cold-stress response & malate accumulation WCI Chr15 (24.8–26.8 Mb) MdMYB73 MD15G1288600 Cold-stress response & malate accumulation IFBD Chr15 (4.6–6.8M b) MdWRKY7 MD15G1078200 Anthocyanin accumulation WCI Chr15 (24.8–26.8 Mb) MdWRKY7 MD15G1287300 Anthocyanin accumulation IFBD Chr15 (43.5–45.5 Mb) MdMYB93 MD15G1369700 Flavonoid & suberin accumulation WCI Chr15 (31.7–34.2 Mb) MdMYB93 MD15G1323500 Flavonoid & suberin accumulation Table 3.
A list of homologues genes/genomic regions associated with the internal flesh browning disorder (IFBD) and red flesh (WCI) in apples. Putative candidate genes residing within these regions are also listed using GDDH13v1.1 reference genome assembly.
-
Mutations in GWAS apple population Predicted gene function Expression in R6 and ‘Royal Gala’ apples Gene Missense
SNPsSNP
StopUpstream
variantsDataset Locus Annotation and TAIR ID Average
RPKM
R6 fleshAverage RPKM
WT fleshlog2Fold
ChangeMD02G1132200 1 FBD Supplemental Table S3 Chr02:9450615-11289014 flavanone 3-hydroxylase
(F3H, TT6, F3'H) AT3G51240387 75 2.47 MD02G1133600 3 FBD Supplemental Table S3 Chr02:9450615-11289014 fatty acid desaturase 5
(FAD5) AT3G15850181 0 9.98 MD02G1153700 1 WCI Table 2 Chr02:11278254-13234556 UDP-Glycosyltransferase, lignin related AT2G18560 1038 534 1.03 MD03G1059200 2 FBD Supplemental Table S3 Chr03:3177994-4910866 Peroxidase AT5G05340 25 1 4.21 MD03G1143300 4 22 FBD Table 1 Chr03:14797661-16611554 bZIP transcription factor (DPBF2, AtbZIP67) AT3G44460 114 852 -2.80 MD03G1147700 12 1 FBD Table 1 Chr03:14797661-16611554 Rho GTPase activating protein AT5G22400 205 89 1.29 MD04G1003400 3 31 WCI Table 2 Chr04:1-1284894 Chalcone synthase (CHS, TT4) AT5G13930 1443 312 2.34 MD04G1204100 6 3 FBD Table 1 Chr04:27629536-29612282 lipoxygenase 1 (LOX1, ATLOX1) AT1G55020 546 268 1.09 MD06G1160700 1 29 FBD Table 1 Chr06:29862341-31737341 peptide met sulfoxide reductase AT4G25130 21212 5456 2.04 MD06G1161400 1 4 FBD Table 1 Chr06:29862341-31737341 Pectin lyase-like protein AT5G63180 6141 29530 -2.22 MD07G1240700 1 26 FBD Supplemental Table S3 Chr07:30357332-31979025 Fe superoxide dismutase 2 AT5G51100 105 1826 -3.99 MD07G1306900 6 FBD Supplemental Table S3 Chr07:34607570-36531467 UDP-glucosyl transferase 78D2 AT5G17050 677 111 2.78 MD08G1249100 2 WCI Supplemental Table S3 Chr08:30486569-31607516 HSP20-like chaperone (ATHSP22.0) AT4G10250 3186 45 6.12 MD09G1114000 3 2 FBD Table 1 Chr09:7999212-11883202 fatty acid desaturase 5 (FAD5) AT3G15850 76 0 8.68 MD09G1146800 1 FBD Table 1 Chr09:7999212-11883202 PHYTOENE SYNTHASE (PSY) AT5G17230 38456 16090 1.32 MD10G1328100 4 5 FBD Supplemental Table S3 Chr10:40235258-41736791 ethylene-forming enzyme (ACO4) AT1G05010 876903 391033 1.21 MD15G1023600 1 FBD Table 1 Chr15:1-1487288 jasmonic acid carboxyl methyltransferase (JMT) AT1G19640 4721 1155 2.07 MD15G1024100 8 FBD Table 1 Chr15:1-1487288 dihydroflavonol 4-reductase (DFR, TT3, M318) AT5G42800 872 309 1.63 MD17G1133400 4 WCI Supplemental Table S3 Chr17:11422073-12433463 PHYTOENE SYNTHASE (PSY) AT5G17230 600 172 1.87 MD17G1260600 2 FBD Supplemental Table S3 Chr17:31098955-32776972 dehydroascorbate reductase 1 (DHAR3) AT5G16710 18 67 −1.78 Table 4.
List of candidate genes associated with WCI or FBD in the XP-GWA and R6:MdMYB10 apple datasets.
Figures
(4)
Tables
(4)