-
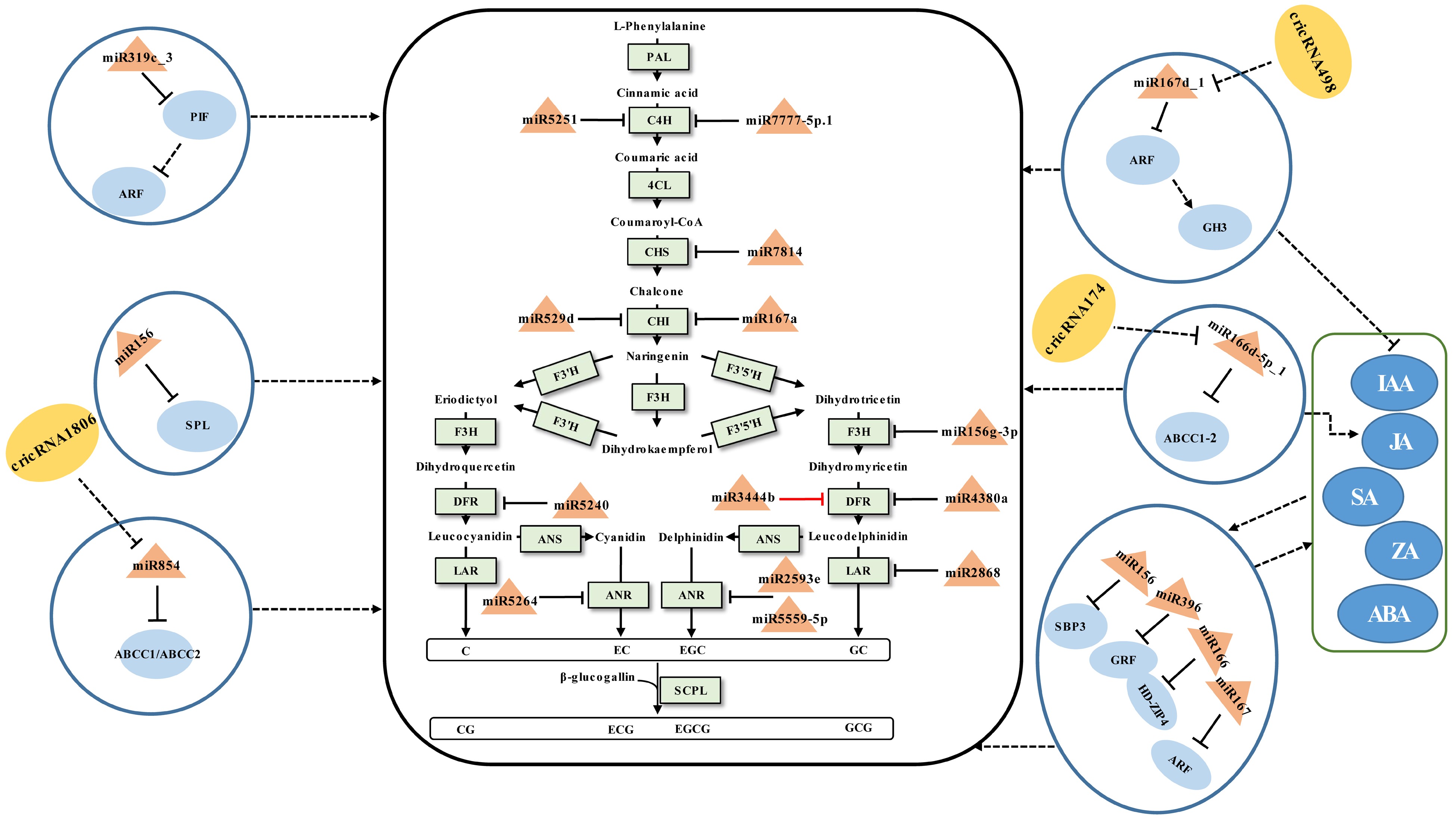
Figure 1.
Schematic representation of the biosynthesis of catechins regulated by ncRNAs in tea plants. The miRNA, circRNA, transcription factor gene, structural gene, and phytohormone are marked in the orange triangles, yellow ovals, light blue ovals, light green boxes, and dark blue ovals, respectively. The solid line with the arrow indicates direct regulation; the dashed line with the arrow is supported by less experimental evidence or prediction results; the black and red T-shaped lines represent negative and positive regulation, respectively; the black dashed T-shaped line is supported by less experimental evidence or prediction results. The miRNA-target pairs shown in the figure have been verified to have regulatory relationships.
-
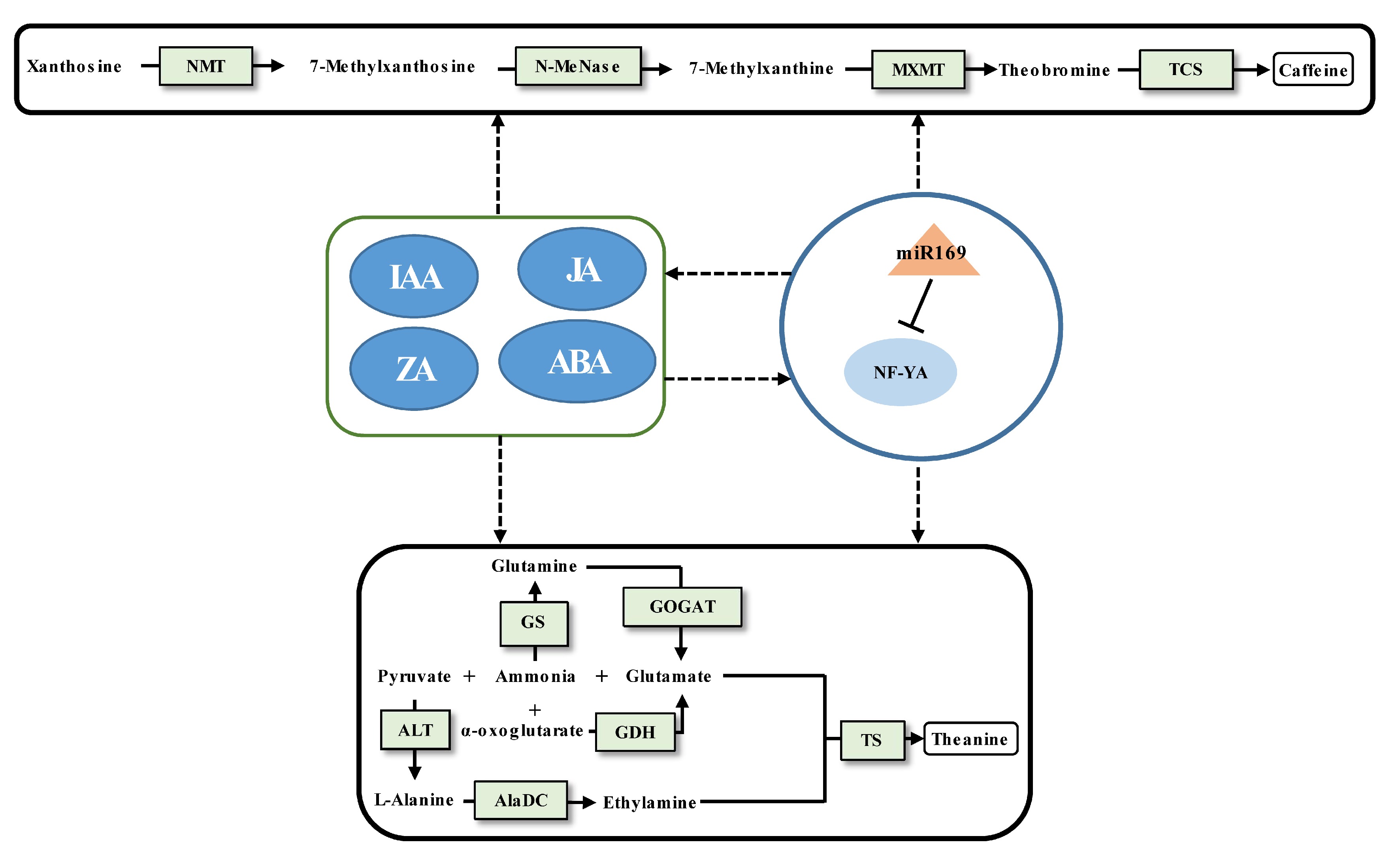
Figure 2.
Schematic representation of the biosynthesis of caffeine and theanine regulated by ncRNAs in tea plants. The miRNA, transcription factor gene, structural gene, and phytohormone are marked by the orange triangle, light blue oval, light green boxes, and dark blue ovals, respectively. The solid line with the arrow indicates direct regulation; the dashed line with the arrow is supported by less experimental evidence or prediction results; and the black T-shaped line represents negative regulation. The miRNA-target pairs shown in the figure have been verified to have regulatory relationships.
-
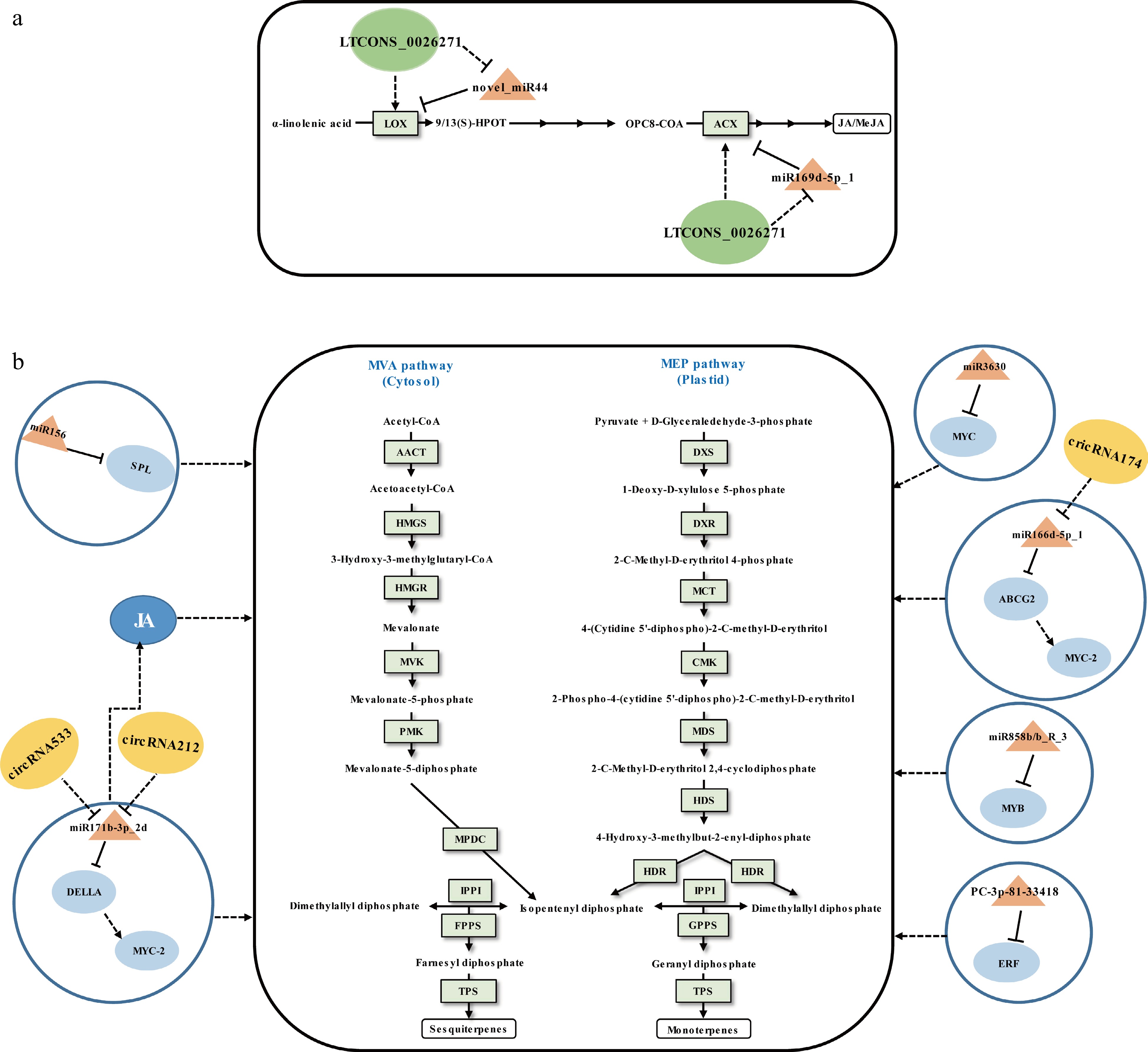
Figure 3.
Schematic representation of the biosynthesis of (a) JA/MeJA and (b) volatile terpenoids in tea plants. The miRNA, circRNA, lncRNA, transcription factor gene, structural gene, and phytohormone are marked in the orange triangles, yellow ovals, green ovals, light blue ovals, light green boxes, and dark blue ovals, respectively. The solid line with the arrow indicates direct regulation; the dashed line with the arrow is supported by less experimental evidence or prediction results; the black T-shaped line indicates negative regulation; the black dashed T-shaped line is supported by less experimental evidence or prediction results. The miRNA-target pairs shown in the figure have been verified to have regulatory relationships.
-
miRNA famliy miRNA name Target Action Verification methods Biology functions Reference miR156 miR156f SPL-3 (Squamosa promoter-binding-like protein) Cleavage 5’RLM-RACE; qRT-PCR Colletotrichum gloeosporioides immune response [59] miR156i SBP (S-RNase binding protein) Cleavage 5’RLM-RACE; qRT-PCR Infection of Pestalotiopsis-like species response [60] miR156 SBP3 Cleavage 5’RLM-RACE; qRT-PCR; northern blot/co-transformation in tobacco leaves Regulation of the biosynthesis of catechins [62] miR156f-3p SBP; AP2/ERF (Ethylene-
responsive transcription factors)Cleavage 5’RLM-RACE; qRT-PCR; northern blot Regulation of the biosynthesis of terpenoids [65] miR156 SPL Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [67] miR156g-3p F3H (flavanone 3-hydroxylase) Cleavage 5’RLM-RACE Regulation of the biosynthesis of catechins [68] miR159 miR159a GAMYB Cleavage 5’RLM-RACE; qRT-PCR The transformation of plant development stages [34] miR160 miR160a-5p ARF17 (Auxin responsive factor17) Cleavage 5’RLM-RACE; qRT-PCR Abiotic stresses response [45] miR160c ARF Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides immune response [47] miR160k Dirigent Cleavage 5’RLM-RACE; qRT-PCR Infection of Pestalotiopsis-like species response [60] miR164 miR164a NAC100 (NAC domain transcription factors100) Cleavage 5’RLM-RACE; qRT-PCR Regulation of leaf development [45] miR164a NAC Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides immune response [47] miR164a NAC-17 Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides immune response [59] miR164a NAC Cleavage 5’RLM-RACE; qRT-PCR; northern blot Regulation of the biosynthesis of catechins [62] miR166 miR166 HD-ZIP III (Homeodomain-
leucine zipperIII)Cleavage 5’RLM-RACE; qRT-PCR Drought stress response [69] miR166a HD-ZIP4 Cleavage 5’RLM-RACE; qRT-PCR; northern blot Regulation of the biosynthesis of catechins [62] miR166d-5p_1 ABCC1-2 (ABC transctipters) Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of flavonoid [61] miR166d-5p_1 ABCG2 Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of terpenoids [61] miR167 miR167a CHL (Chalcone isomerase) Cleavage 5’RLM-RACE; qRT-PCR Regulation of flavonoid biosynthesis [44] miR167a ARF6 Cleavage 5’RLM-RACE Regulation of the biosynthesis of catechins [44] miR167a ARF8 Cleavage 5’RLM-RACE Regulation of the biosynthesis of catechins [44] miR167 ARF Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [34] miR167d_1 ARF Cleavage 5’RLM-RACE; qRT-PCR; miRNA-agomir/antagomir Regulation of the biosynthesis of flavonoid and terpenoids [61] miR167d_1 GH3 (Flavanone 3-hydroxylase) Cleavage qRT-PCR; miRNA-agomir/antagomir Regulation of the biosynthesis of flavonoid and terpenoids [61] miR167d ARF Cleavage 5’RLM-RACE; qRT-PCR; northern blot Regulation of the biosynthesis of catechins [62] miR169 miR169e NFY (Nuclear transcription
factor Y)Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides immune response [47] miR169d-5p_1 ACX (acyl-CoA oxidase) Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of JA/MeJA and terpenoids [70] miR169a NF-YA (Nuclear
factor Y)Cleavage 5’RLM-RACE; qRT-PCR; northern blot Regulation of the biosynthesis of theanine and caffeine [62] miR171 miR171b-3p CRE1 (cytokinin receptor 1) Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of flavonoid and terpenoids [61] miR171b-3p_2 DELLA1 (DELLA protein1) Cleavage 5’RLM-RACE; qRT-PCR; miRNA-agomir/antagomir Regulation of the biosynthesis of flavonoid and terpenoids [61] miR172 miR172k ERF_RAP2-7 Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides immune response [59] miR172g-3p MYC2 Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of terpenoids [61] miR319 miR319c TPC (TCP family transcription factor) Cleavage 5’RLM-RACE C. gloeosporioides immune response [59] miR395 miR395e APS (Aspartic proteases) Cleavage 5’RLM-RACE; qRT-PCR Infection of Pestalotiopsis-like species response [60] miR396 miR396b-5p GRF-1 Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides immune response [59] miR396b-5P F3H Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of flavonoid [61] miR396 GRF Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [34] miR396d GRF1 Cleavage 5’RLM-RACE; qRT-PCR; northern blot Regulation of the biosynthesis of catechins [62] miR396d GRF4 Cleavage 5’RLM-RACE; qRT-PCR; northern blot; co-transformation Regulation of the biosynthesis of catechins [62] miR396d GRF13 Cleavage 5’RLM-RACE; qRT-PCR; northern blot Regulation of the biosynthesis of catechins [62] miR397 miR397 LAC17 (Laccase17) Cleavage 5’RLM-RACE; qRT-PCR Infection of Pestalotiopsis-like species response [60] miR398 miR398a-3p-1 CSD4 (Cu/Zn-Superoxide dismutases4) Cleavage 5’RLM-RACE; qRT-PCR Cold stress response [66] miR408 miR408-3p_2ss18GT19GT STK1 (Ser/Thr-protein Kinse) Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides immune response [47] miR447 miR447g-p5 PAL (Phenylalanine ammonia-lyase) Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides immune response [59] miR529 miR529d CHI (chalcone isomerase) Cleavage 5’RLM-RACE; Regulation of the biosynthesis of catechins [68] miR530 miR530b ERF96 Cleavage 5’RLM-RACE; qRT-PCR; northern blot/co-transformation Infection of Pestalotiopsis-like species response [60] miR530a DHBP Cleavage 5’RLM-RACE; qRT-PCR Infection of Pestalotiopsis-like species response [60] miR828 miR828 WER Cleavage 5’RLM-RACE; qRT-PCR Regulation of plant development [45] miR828a MYB (MYB domain protein) Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of terpenoids [65] miR828a MYB75 Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides immune response [47] miR845 miR845 ABCC1-3 Cleavage 5’RLM-RACE; qRT-PCR/miRNA-agomir/antagomir Regulation of the biosynthesis of flavonoid and terpenoids [61] miR845 ABCC2 Cleavage qRT-PCR; miRNA-agomir/antagomir Inhibite flavonoid biosynthesis but enhance terpenoid biosynthesis [61] miR858 miR858a MYB12 Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of polyphenols [45] miR858b_R-3 MYB Cleavage 5’RLM-RACE; qRT-PCR; northern blot Regulation of the biosynthesis of terpenoids [65] miR858b MYB Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of terpenoids [65] miR858a R2R3-MYB Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides immune response [59] 858b_R-3 MYB114 Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides immune response [47] miR2593 miR2593e ANR (Anthocyanidinreductase) Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [44] miR2868 miR2868 LAR (leucoanthocyanidin reductase) Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [68] miR3444 miR3444b DFR (Dihydroflavonol 4-reductase) Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [44] miR3630 miR3630-3p_L-3p MYC Cleavage qRT-PCR; northern blot Regulation of the biosynthesis of terpenoids [65] miR4380 miR4380a DFR Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [62] miR5240 miR5240 DFR Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [68] miR5251 miR5251 C4H Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [62] miR5559 miR5559-5p ANR1; ANR2 Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [68] miR5564 miR5564 ANR2 Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [68] miR7777 miR7777-5p C4H Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [62] miR7814 miR7814 CHS1 (chalcone synthase1) Cleavage 5’RLM-RACE Regulation of the biosynthesis of catechins [68] miR7814 CHS3 Cleavage 5’RLM-RACE Regulation of the biosynthesis of catechins [68] miR8757 miR8757f PPO (Polyphenol oxidase) Cleavage 5’RLM-RACE; qRT-PCR Infection of Pestalotiopsis-like species response [60] novel miRNA miRn211 TLP (thaumatin-like protein) Cleavage 5’RLM-RACE; qRT-PCR; northern blot; co-transformation Infection of Pestalotiopsis-like species response [60] PC-5p-80764_22 WRKY (WRKY transcription
factor)Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides and abiotic stresses response [47] PC-3p-81-33418 AP2/ERF Cleavage qRT-PCR; northern blot Regulation of the biosynthesis of terpenoids [65] novel_miR44 LOX (Lipoxygenase) Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of JA/MeJA and terpenoids [70] Table 1.
Summary of validated miRNA-target pairs in tea plant.
Figures
(3)
Tables
(1)