-

Figure 1.
Phenotypes of VwTYDC-silenced Viola × wittrockiana. (a) Blank control; (b) flower phenotype after infection with pTRV2-VwCHS; (c) flower phenotype after infection with pTRV2-VwANS; (d) flower phenotype after infection with pTRV2-VwTYDC.
-
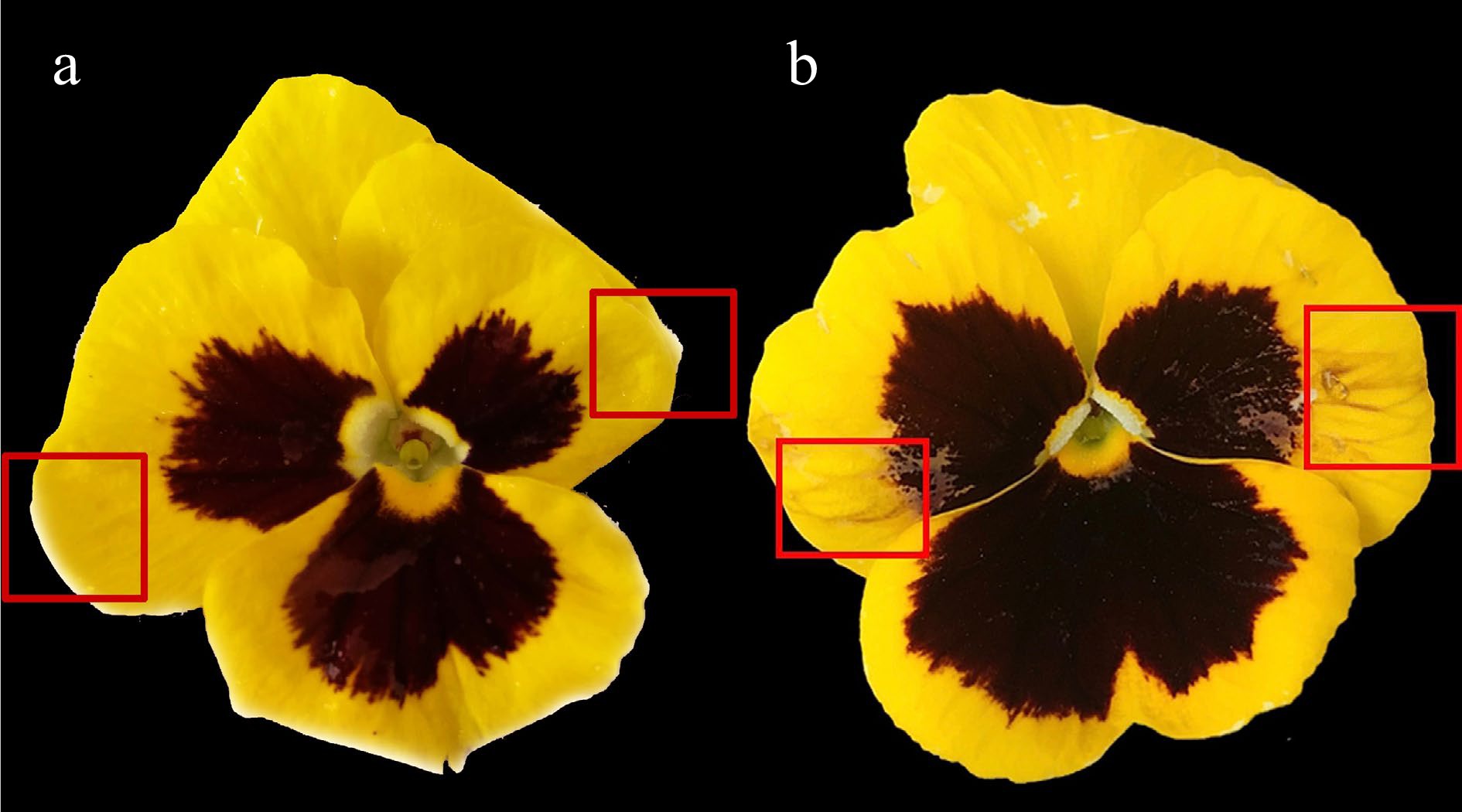
Figure 2.
Effect of tyrosine treatment on non-blotched areas of Viola × wittrockiana petals. (a) ddH2O treatment; (b) tyrosine treatment. The treated areas are marked by a red box.
-

Figure 3.
Violin plot of anthocyanins in petals of Viola × wittrockiana treated with tyrosine or ddH2O. The box in the middle represents the interquartile range. The black horizontal line in the middle is the median, and the outer shape represents the distribution of the data.
-
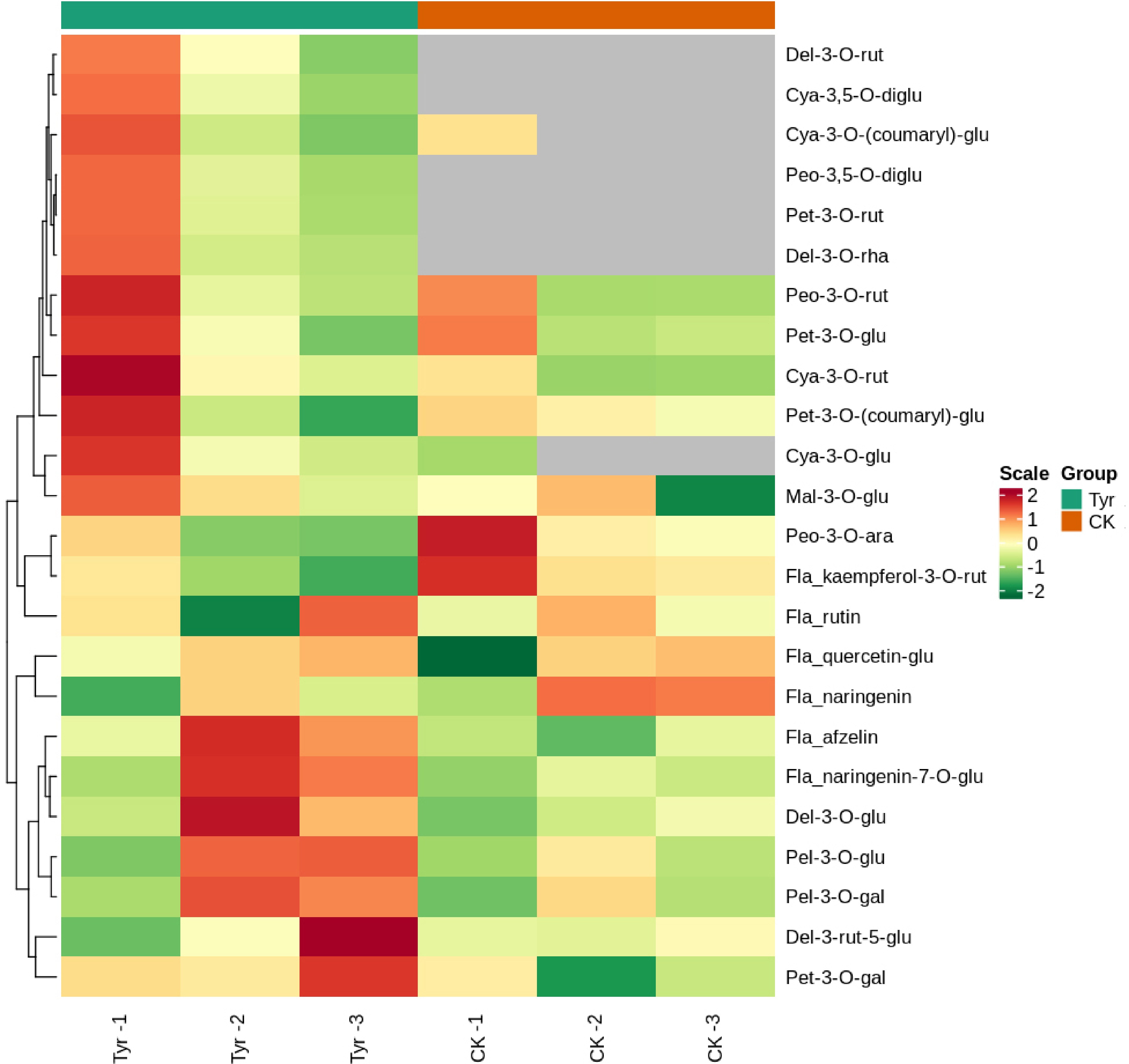
Figure 4.
Heatmap of the contents of the 24 compounds detected in different Viola × wittrockiana samples analyzed by hierarchical clustering analysis. The horizontal axis shows the sample information, and the vertical axis shows the metabolite information. The tree on the left of the figure shows the metabolite clustering, and the scale is the metabolite content after standardization. A deeper red color indicates a higher content, and gray indicates that the compound was not detected. Groups indicate the different treatments. CK: ddH2O treatment; Tyr: tyrosine treatment.
-
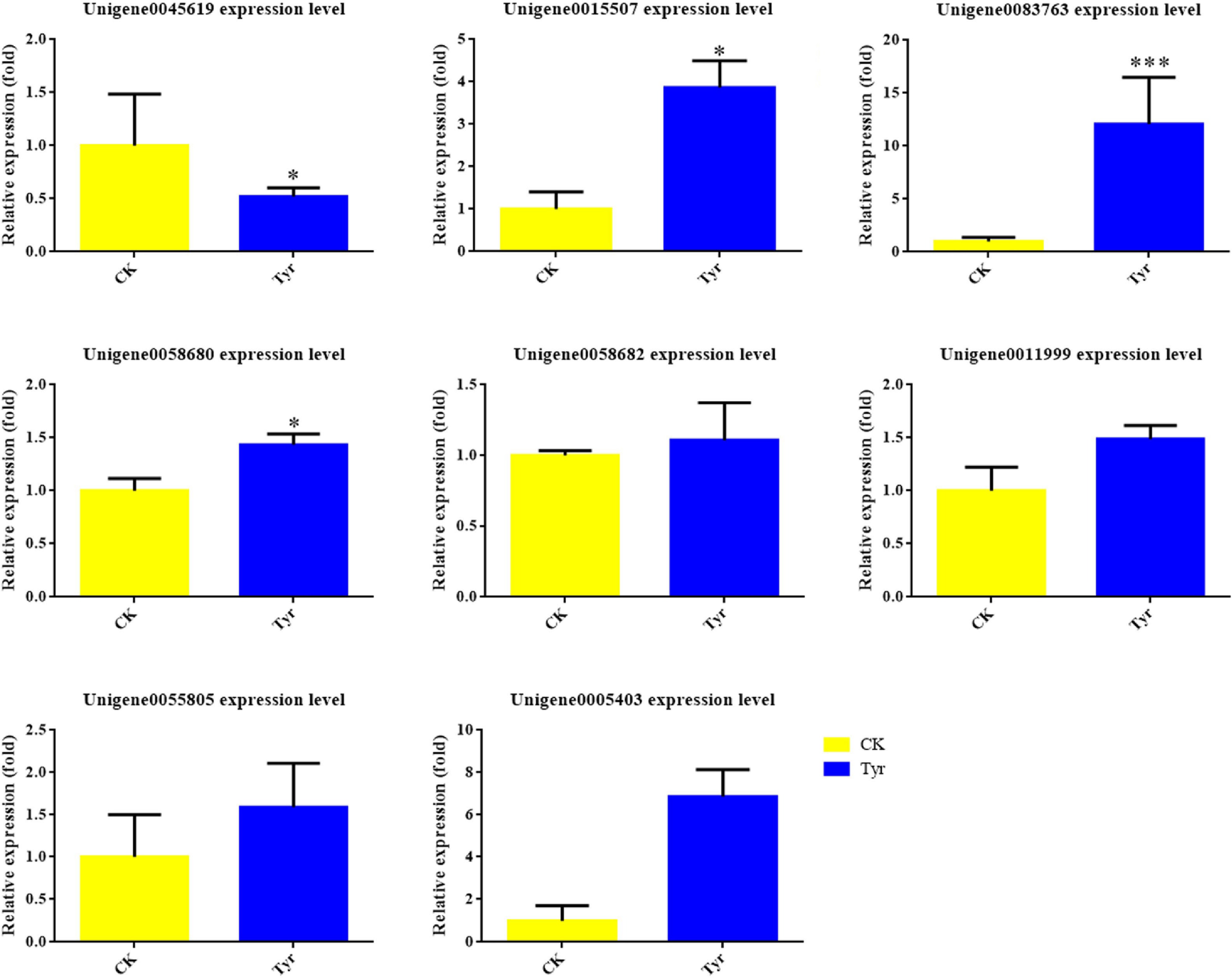
Figure 5.
The relative expression levels of anthocyanin biosynthesis and transcription factor genes in Viola × wittrockiana petals that received tyrosine (Tyr) or ddH2O (CK) treatment. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001.
-
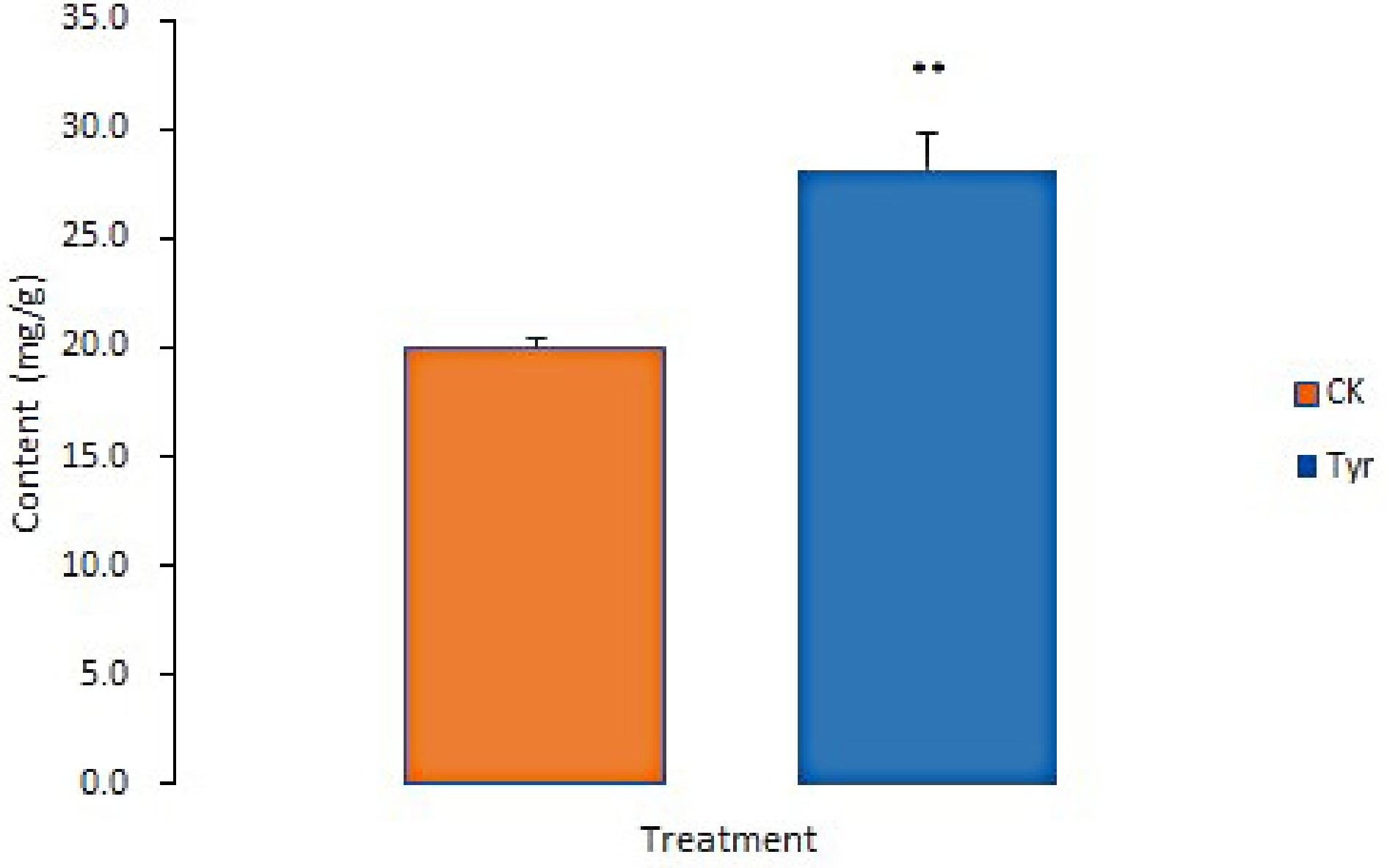
Figure 6.
ABA contents in Viola × wittrockiana from different treatment groups. ** p ≤ 0.01. CK, ddH2O treatment. Tyr, tyrosine treatment.
-
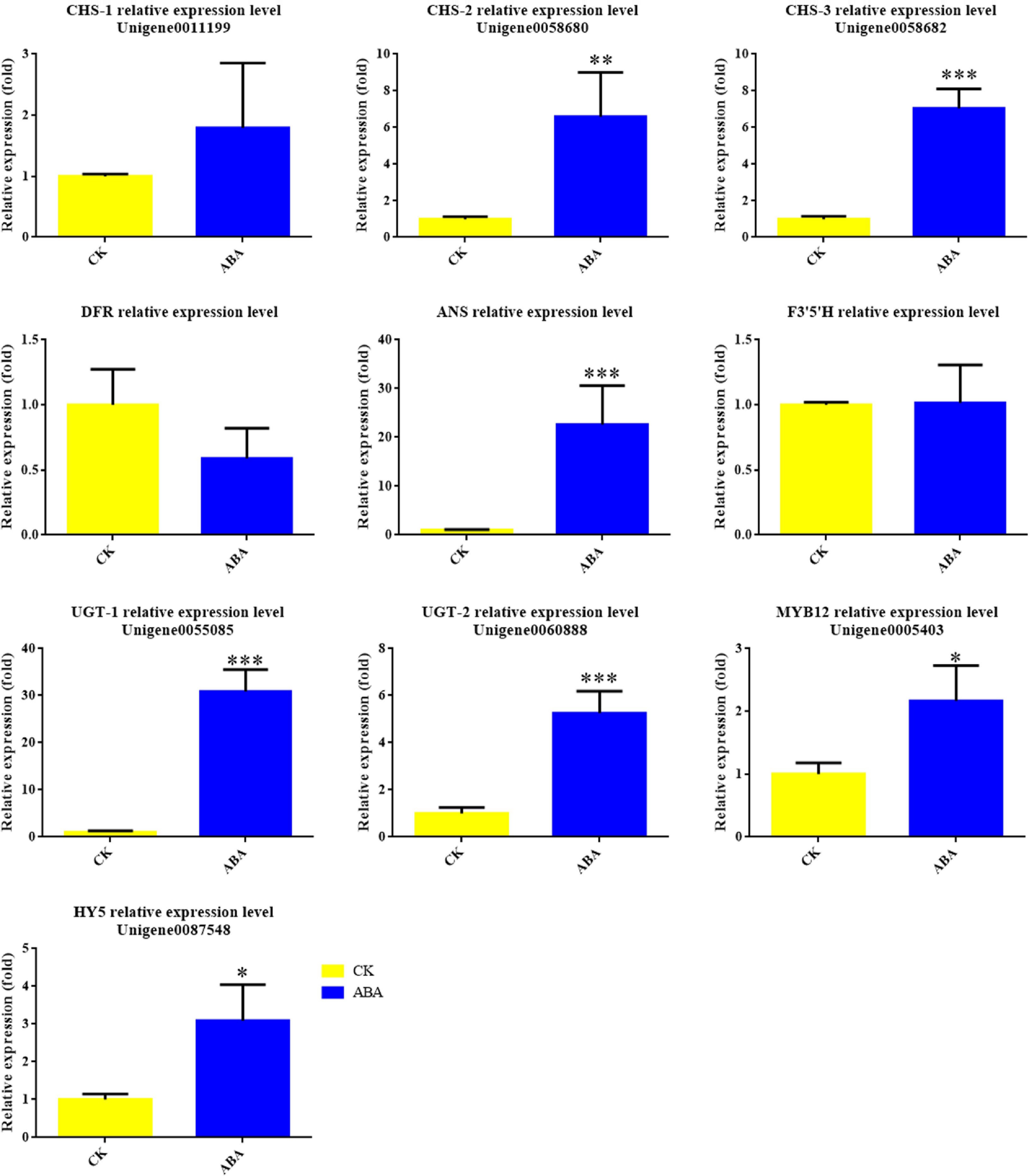
Figure 7.
Expression levels of anthocyanin biosynthesis genes and transcription factors in Viola × wittrockiana with and without ABA treatment. * p ≤ 0.05, ** p ≤ 0.01, ***p ≤ 0.001.
-
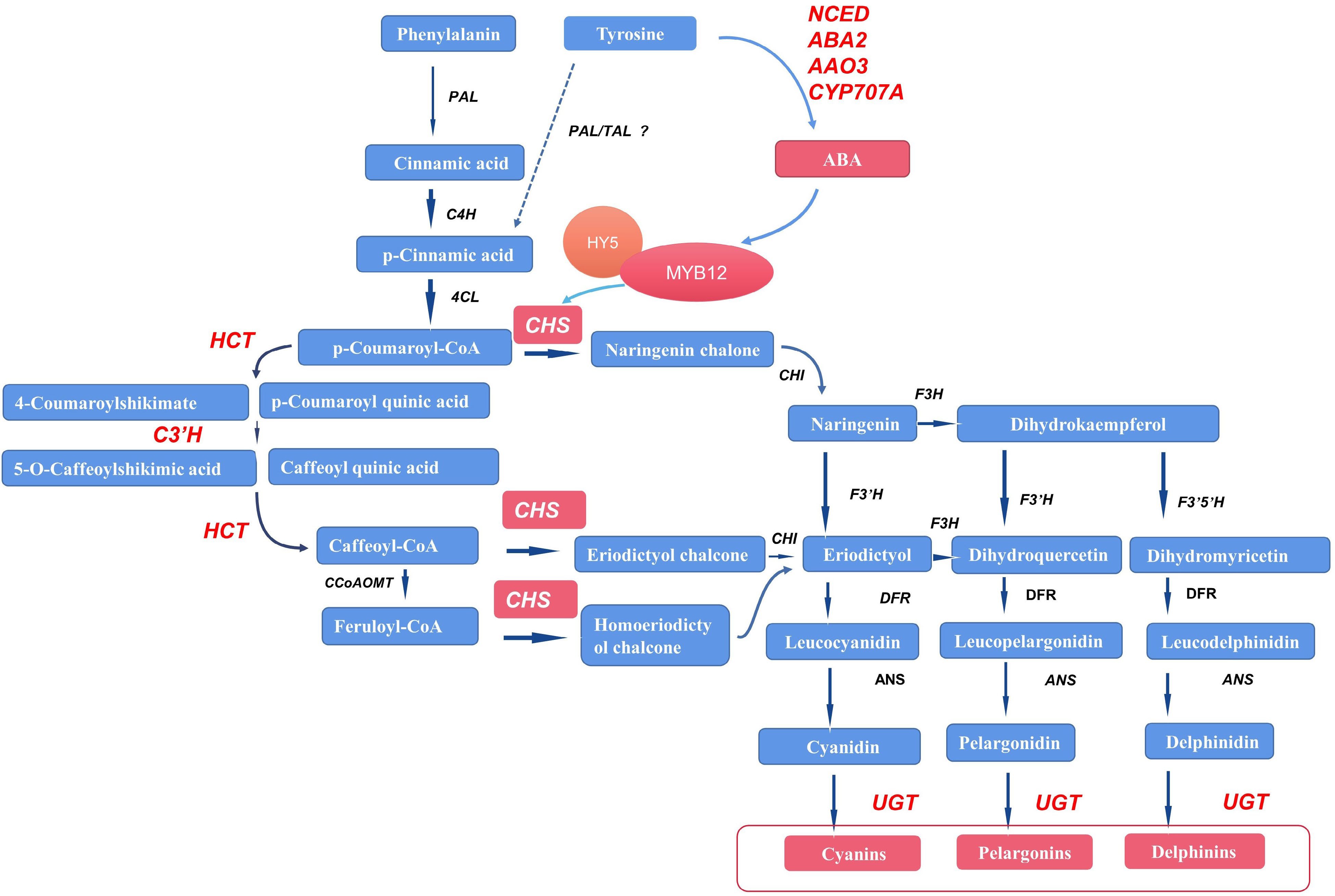
Figure 8.
Possible pathways by which tyrosine treatment may promote anthocyanin biosynthesis in Viola × wittrockiana. The red color represents upregulation of genes and metabolites. The dashed arrow represents the possible pathway. PAL/TAL indicates that PAL may function like TAL in pansy, although this remains to be verified. PAL: phenylalanine ammonia-lyase; TAL: tyrosine ammonia-lyase; C4H: cinnamate 4-hydroxylase; 4CL: 4-coumarate-CoA ligase; HCT: shikimate O-hydroxycinnamoyltransferase; C3′H: 5-O-(4-coumaroyl)-D-quinate 3'-monooxygenase; CHS: chalcone synthase; F3H: naringenin 3-dioxygenase; F3′H: flavonoid 3'-hydroxylase; F3′5′H: flavonoid 3',5'-hydroxylase; DFR: flavanone 4-reductase; ANS: anthocyanidin synthase; UGT: UDP glucuronosyltransferase, falconoid 3-O-glycosyltransferase; CCoAOMT: caffeoyl-CoA O-methyltransferase; VwNCED: 9-cis-epoxycarotenoid dioxygenase; VwABA2: xanthoxin dehydrogenase; VwAAO3: abscisic-aldehyde oxidase; VwCYP707A: (+)-abscisic acid 8'-hydroxylase.
-
Gene ID Annotation RPKM
(Tyr treatment)RPKM
(H2O treatment)log2 (T/CK) FDR Up/Down unigene
0045619VwHCT 0.54 1.48 −1.45 0.05 Down unigene
0015507VwHCT 2.31 0.56 2.26 5.35e-12 Up unigene
0083763VwC3’H 3.60 0.34 3.42 1.18e-28 Up unigene
0029000VwC3’H 1.80 0.02 6.86 8.38e-15 Up unigene
0058680VwCHS 213.61 100.30 1.09 4.65e-35 Up unigene
0058682VwCHS 87.86 42.41 1.05 1.18e-41 Up unigene
0011199VwCHS 10.51 3.40 1.63 6.24e-33 Up unigene
0060888VwUGT75C1 13.30 5.71 1.22 2.95e-21 Up unigene
0055085VwUGT75C1 177.86 66.99 1.41 3.19e-105 Up VwHCT, shikimate O-hydroxycinnamoyltransferase; VwC3'H, coumaroylquinate 3'-monooxygenase; VwCHS, chalcone synthase; VwUGT75C1, anthocyanidin 3-O-glucoside 5-O-glucosyltransferase. Table 1.
Putative anthocyanin structural genes that were differentially expressed in response to tyrosine treatment in Viola × wittrockiana petals.
-
Gene ID Annotation RPKM
(Tyr treatment)RPKM
(H2O treatment)log2
(T/CK)FDR Up/Down unigene
0004917VwNCED 2.20 1.03 1.10 0.00 Up unigene
0068071VwNCED 2.41 0.56 2.10 7.18e-6 Up unigene
0068072VwNCED 1.16 0.35 1.72 0.00 Up unigene
0082413VwABA2 0.33 0.01 4.51 0.00 Up unigene
0073280VwAAO3 8.81 4.28 1.04 2.46e-31 Up unigene
0017204VwCYP707A 18.30 0.81 4.50 7.13e-212 Up unigene
0087548VwHY5 0.63 0 9.29 0.01 Up VwNCED, 9-cis-epoxycarotenoid dioxygenase; VwABA2, xanthoxin dehydrogenase 2; VwAAO3, abscisic-aldehyde oxidase; VwCYP707A, (+)-abscisic acid 8'-hydroxylase. Table 2.
Differentially expressed genes related to ABA biosynthesis in Viola × wittrockiana.
Figures
(8)
Tables
(2)