-
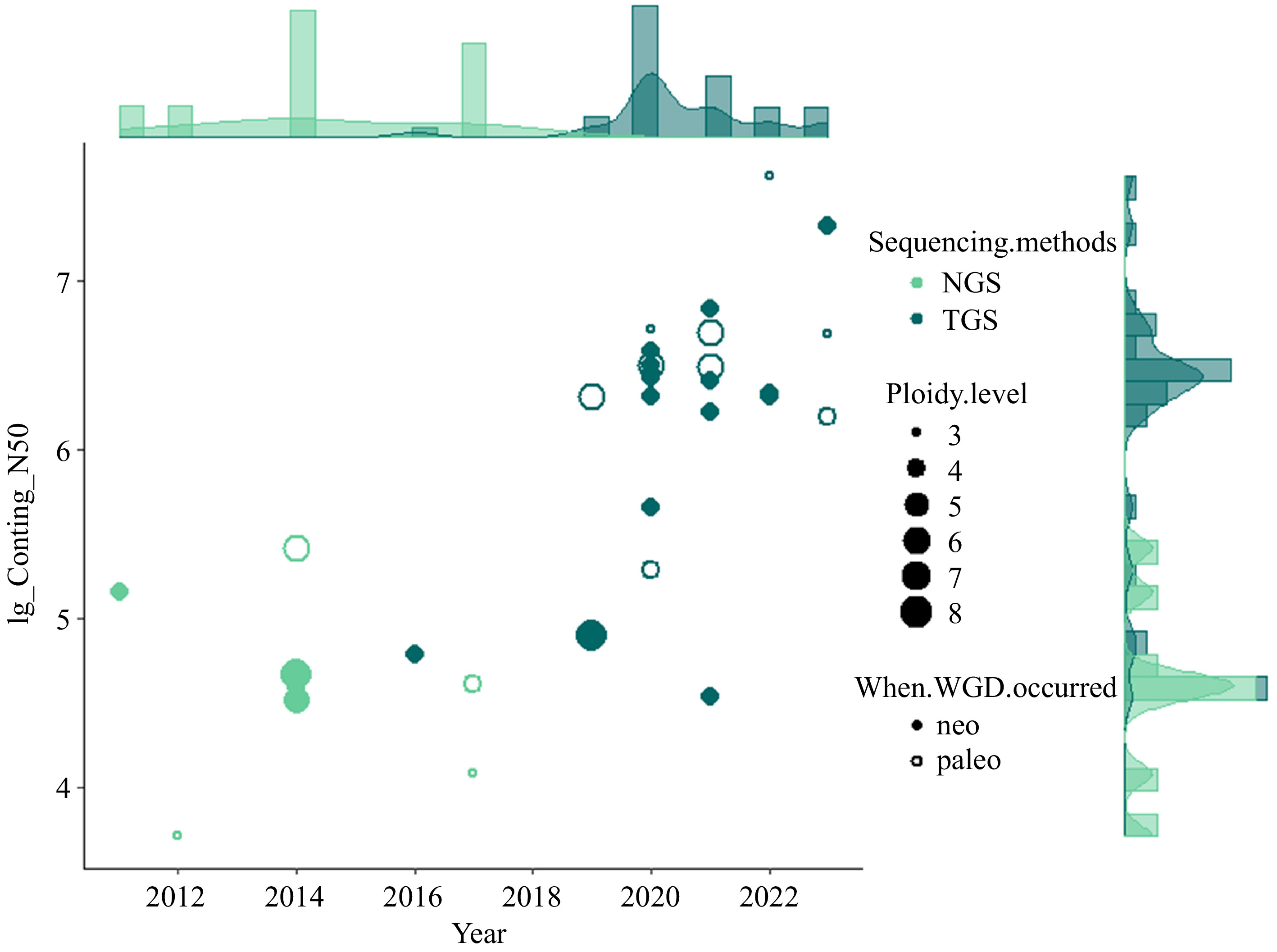
Figure 1.
Overview of currently sequenced polyploid vegetables with the contig N50 (log10) of assemblies and publication year as Y and X axis, respectively. The type of sequencing technology (Next-generation sequencing, NGS and Third-generation sequencing, TGS) are indicated with different colors. The size of data points is scaled by the ploidy level. Neo- and paleo- polyploids are differentiated with closed and open data points.
-
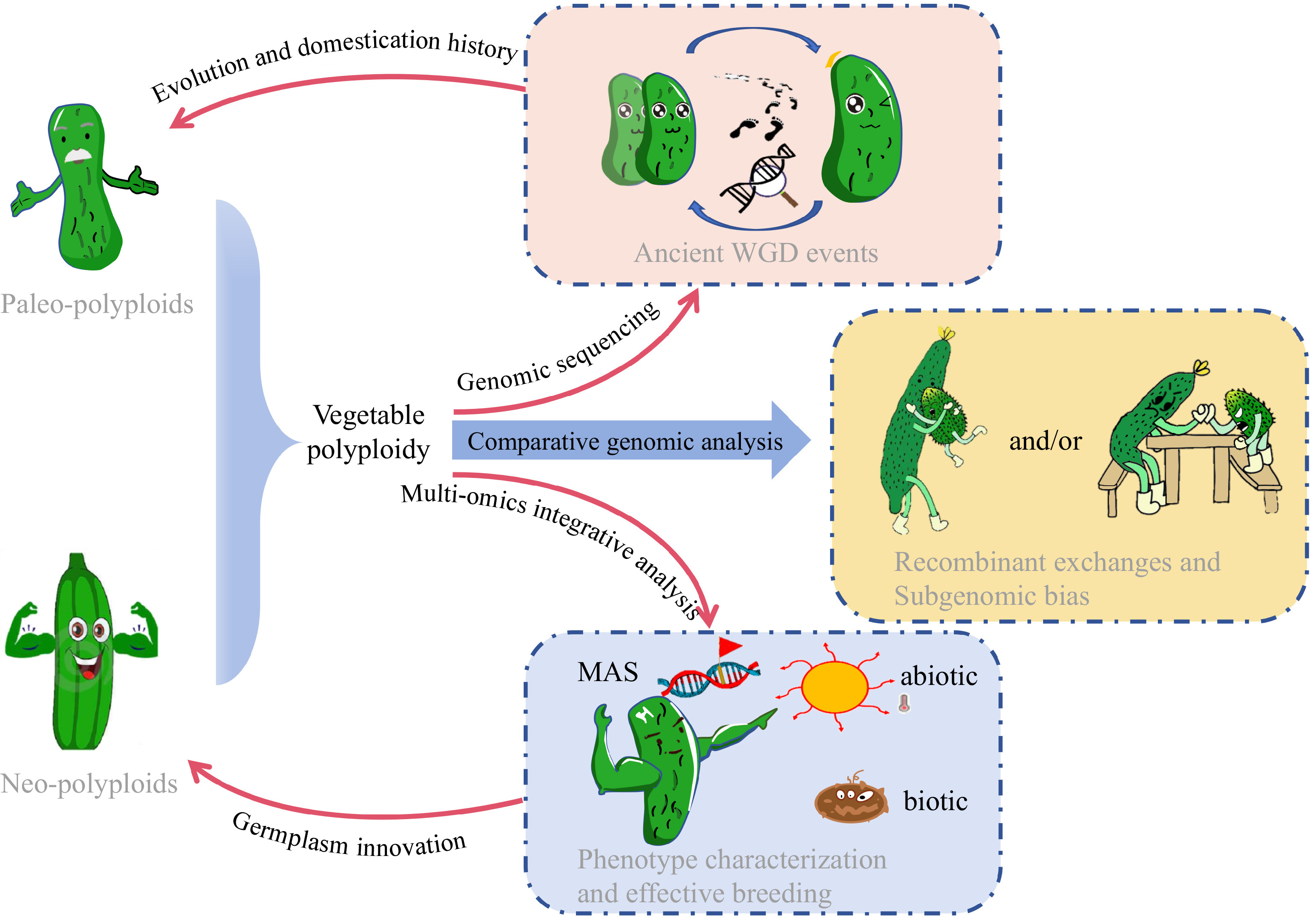
Figure 2.
Schematic diagram illustrating the application of high-throughput sequencing for evolutionary genomics, promoting germplasm innovation and effective breeding in neo- and paleo- polyploid vegetables with cucumber as an example.
-
Species Ref Analysis tools Year of publication Solanum tuberosum [38] SOAPdenovo; EMBOSS; SOAPdenovoalign 2011 Solanum lycopersicum [39] Newbler; CABOG; BAC FISH; EuGene; ABySS 2012 Brassica napus [40] SOAPdenovo; Newbler; GapCloser; 2014 Fragaria × ananassa [25] SOAPdenovo; Newbler; GapCloser; 2014 Camelina sativa [41] SOAPdenovo; Bambus; GapCloser; NUCmer 2014 Brassica oleracea [42] SOAPdenovo; Bambus; GapCloser; 2014 Brassica juncea var. tumida [43] ALLPATHS-LG; PBjelly; IrysView 2016 Cucurbita maxima Duch. [44] ShortRead; QuorUM; SOAPdenovo; Newbler; GapCloser; Pilon 2017 Lactuca sativa [45] SOAPdenovo; HiRis 2017 Fragaria × ananassa [46] NRGene; HiRise; PBJelly; Pilon; CoGe 2019 Brassica oleracea [47] FALCON; Quiver; BWA-MEM 2019 Brassica napus [48] Falcon; CANU; pbalign; pilon; 3D-DNA; Juicebox; Juicer 2020 Medicago sativa [49] CANU; MECAT; HERA; BWA-MEM; Redundans; Purge Haplotigs; BWA-MEM; Pilon 2020 Medicago sativa [36] CANU; ALLHiC; Jcvi; Juicebox 2020 Brassica oleracea [50] PBcR wgs8.3rc1; FALCON; Quiver; Pilon; BWA-MEM; SALSA; Mummer 2020 Solanum melongena [51] wtdbg2; Racon; Pilon; fragScaff; LACHESIS 2020 Allium sativum [52] FALCON; Quiver; Pilon; Purge Haplotigs; FragScaff; Burrows-Wheeler Aligner; ALLHiC 2020 Brassica napus [53] CANU; Pilon; 3D-DNA; ALLMAPS; ALLHiC 2021 Brassica juncea [54] CANU; BWA mem; MaSuRcA; RefAligner; IrysSolve; Pilon 2021 Brassica juncea [55] Jellyfish; FALCON; Sspace-longread; Quiver; BioNano Solve; BioNano Access; Juicebox; HiCPlotter 2021 Cucumis × hytivus [26] CANU; Pilon; SOAPdenovo; SSPACE; RefAligner; cutadapt; LACHESIS; HiC-Pro 2021 Brassica oleracea [56] Falcon; Arrow; PBJelly 2021 Solanum tuberosum [22] pbccs; hifiasm; BWA-MEM; ALLHiC; jcvi 2022 Solanum tuberosum [57] hifiasm; pilon; 2022 Capsicum annuum [58] MECAT2; Pilon; CANU; Juicer; Juicerbox; 3D-DNA; Phred; CA59 2022 Raphanobrassica [59] JELLYFISH; CANU; Arrow; Pilon; JUICER; 2023 Vicia faba [60] hifiasm; Sniffles; findGSE; purge_haplotigs; Merqury; RagTag; GSAlign; Liftoff 2023 Lactuca sativa [61] CANU; WTDBG2; Arrow; Pilon; 3D-DNA; Juicer; Falcon 2023 Table 1.
Analysis tools used for vegetable polyploid genome assembly.
Figures
(2)
Tables
(1)