-

Figure 1.
Symptoms of cassava brown streak disease. (a) Leaf chlorosis without distortion of the lamina and brown lesions on a green stem. (b) Storage root constrictions. (c) Mild necrosis of cassava storage roots. (d) Severe necrosis of cassava storage roots.
-
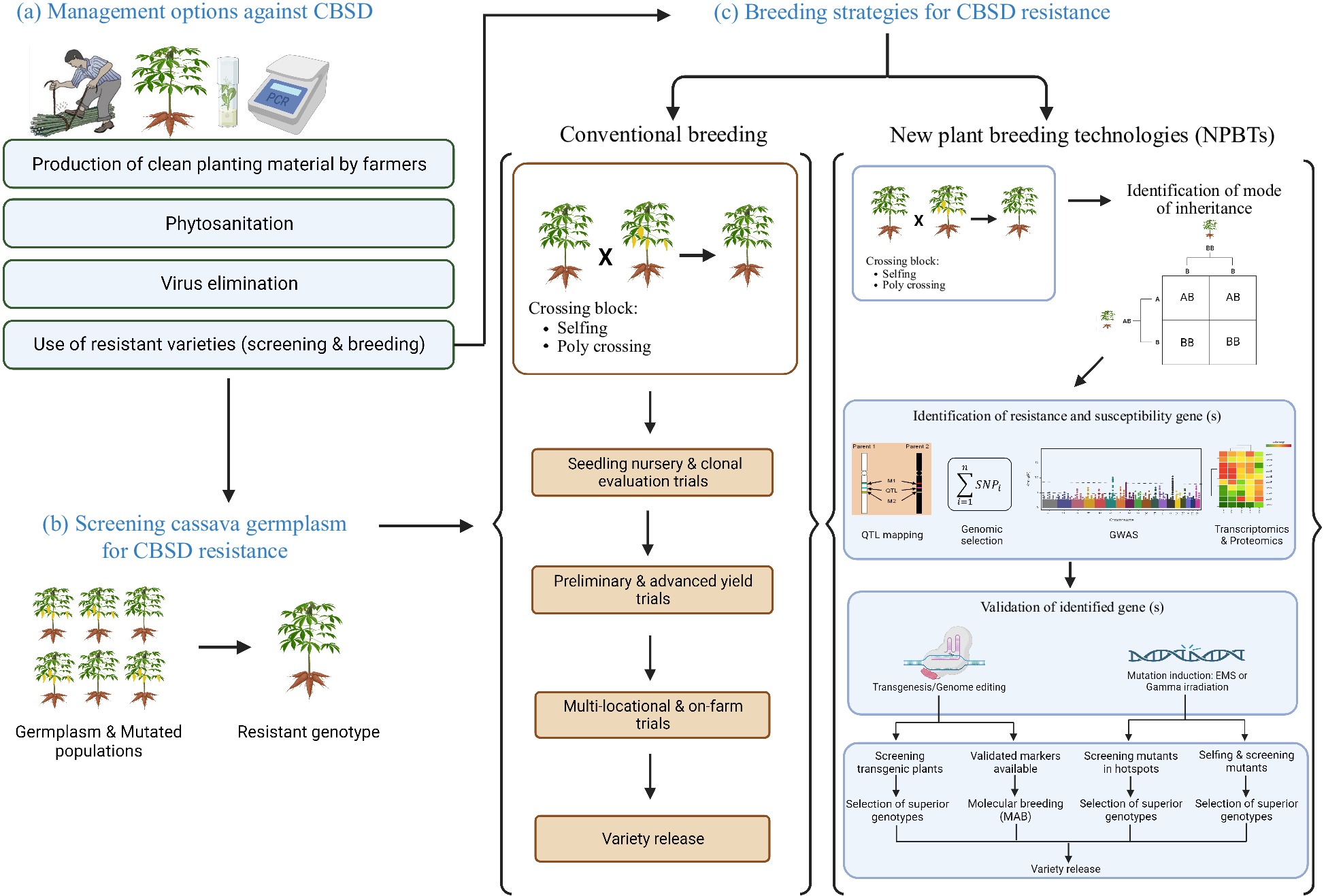
Figure 2.
Illustration of management and breeding strategies used to mitigate CBSD in Africa. (a) Various CBSD management options were used. Those are the production of clean planting material by farmers, phytosanitation, virus elimination using tissue culture, thermotherapy and chemotherapy and the use of resistant varieties. (b) Screening cassava germplasm and mutated population for CBSD resistance can be carried out either in the fields (CBSD hotspots) or in controlled environments where grafting method or infectious clones can be used. (c) In conventional breeding, a resistant genotype is crossed with a susceptible one and GCA and SCA for CBSD resistance are evaluated in progenies. Various genomic tools and NPBTs for CBSD resistance were used. Crossing of an identified resistant parent with susceptible farmer-preferred genotype is followed by a determination of the mode of inheritance, the candidate genes, and then validation of candidate genes. Validated markers are used as inputs in molecular breeding. After gene validation, field experiments are conducted to screen and select adapted superior genotypes for release. Conventional breeding is lengthy because it can take 8−10 years. Using NPBTs can help to shorten this cycle to few years: starting with mapping population development for marker discovery (2 years), marker development and validation (1 year) and field trials (2 years), the breeding cycle can take about 5 years. Once the markers are available, this cycle can be even shorter i.e., 3−4 years. This figure was created with BioRender.com.
-
Table 1.
List of countries where cassava brown streak disease is reported.
-
No Objective of the study Key findings Reference 1 To identify molecular markers associated with resistance against CBSD Two consistent QTLs were associated with CBSD root necrosis resistance on chromosomes II and XI, plus a putative QTL located on chromosome XVIII. [116] 2 To identify QTL associated with resistance to CBSD root necrosis and CBSD foliar symptoms Two QTLs were associated with only CBSD root necrosis (on chromosomes V and XII), seven QTLs were associated with only CBSD foliar symptoms (detected on chromosomes IV, VI, XVII and XVIII) and two QTLs that were associated with both CBSD foliar and root necrosis on chromosomes XI and XV. [117] 3 To map QTL associated with resistance to foliar and root necrosis CBSD symptoms Two QTLs (1 minor and 1 major) for foliar CBSD symptoms on chromosome XI and XVIII. The major QTL for foliar symptoms explained 12.87% of PHV at 6 MAP. [119] 4 To determine CBSD resistance-associated allele frequency and distribution and investigate the genetic relationships between some CBSD resistant genotypes A low allele frequency was putatively associated with Namikonga derived resistance to CBSD and there was a wide distribution of this frequency in the south-eastern and central African germplasm. [120] 5 To evaluate the potential of GS to enhance CBSD resistance via assessing the accuracy of 7 genomic prediction models Results showed good predictive ability of 0.40–0.42 for foliar CBSD severity and 0.31–0.42 for CBSD root severity at 6 MAP. [132] 6 To assess the use of genomic predictions of West African clones by using training data from a Ugandan population Higher genomic prediction accuracies for CBSD resistance were obtained by using optimized East African training populations. [133] 7 To assess the efficacy of GS for improving cassava for CBSD resistance and other traits Low predictive abilities for CBSD resistance (mean of 0.20 for
G-BLUP) were reported.[135] 8 To evaluate the association of 5 eIFs with cassava tolerance and susceptibility responses to CBSD Results showed that two SNPs in two genes were weakly associated with the CBSD but without any direct causal-effect relationship. [136] 9 To examine the power of diverse germplasm assembled
from two cassava breeding programs in Tanzania at
different breeding stages to predict traits and discover
quantitative trait loci (QTL)QTLs associated with CBSD resistance were found on chromosomes 9 and 11 while QTLs detected on chromosomes 2, 3, 8 and 10 were associated with resistance to CBSD root necrosis. Other three QTLs for dual resistance to CMD and CBSD were detected on chromosome 4 (one QTL) and chromosome 12 (2 QTLs). [118] 10 To generate eIF isoform (nCBP1,nCBP2 and nCBP1/nCBP2) mutants in line 60444 and assess their reponses upon challenge with CBSV Results showed that ncbp-1/ncbp-2 mutants exhibited not only delayed and attenuated CBSD aerial symptoms, but also reduced severity and incidence of storage root necrosis. [137] CBSD: Cassava brown streak disease; CBSV: cassava brown streak virus; QTL: Quantitative trait locus; GS: Genomic selection; MAP: Months after planting; SNP: Single nucleotide polymorphism; eIF: Eukaryotic translation initiation factors. Table 2.
Summary of published studies on NPBTs towards CBSD management.
Figures
(2)
Tables
(2)