-
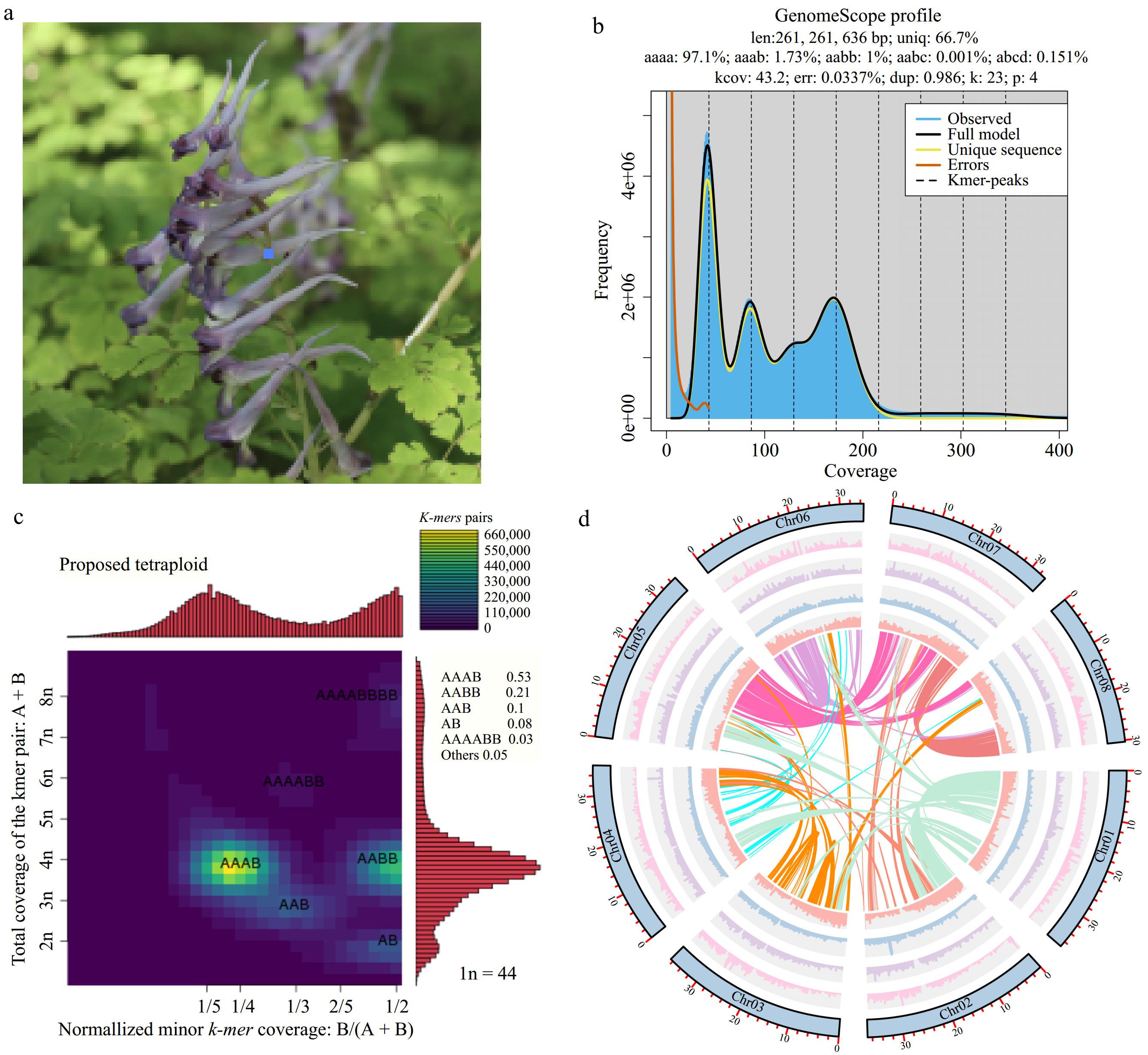
Figure 1.
Overview of Corydalis sheareri genome. (a) Photo of C. sheareri. (b) Genome survey of C. sheareri. (c) Heatmap for coverage pattern of heterozygous k-mer pairs, in which X axis indicates the normalized minor k-mer coverage, while Y axis indicates the total k-mer pairs coverage. (d) Genomic features of eight pseudochromosome. The outermost circle (blue) represents each chromosome of the genome. The bar charts of the second to fifth circles suggest gene density, LTR density, Copia, and Gypsy density, respectively. The inner circular shows inter-chromosomal synteny.
-
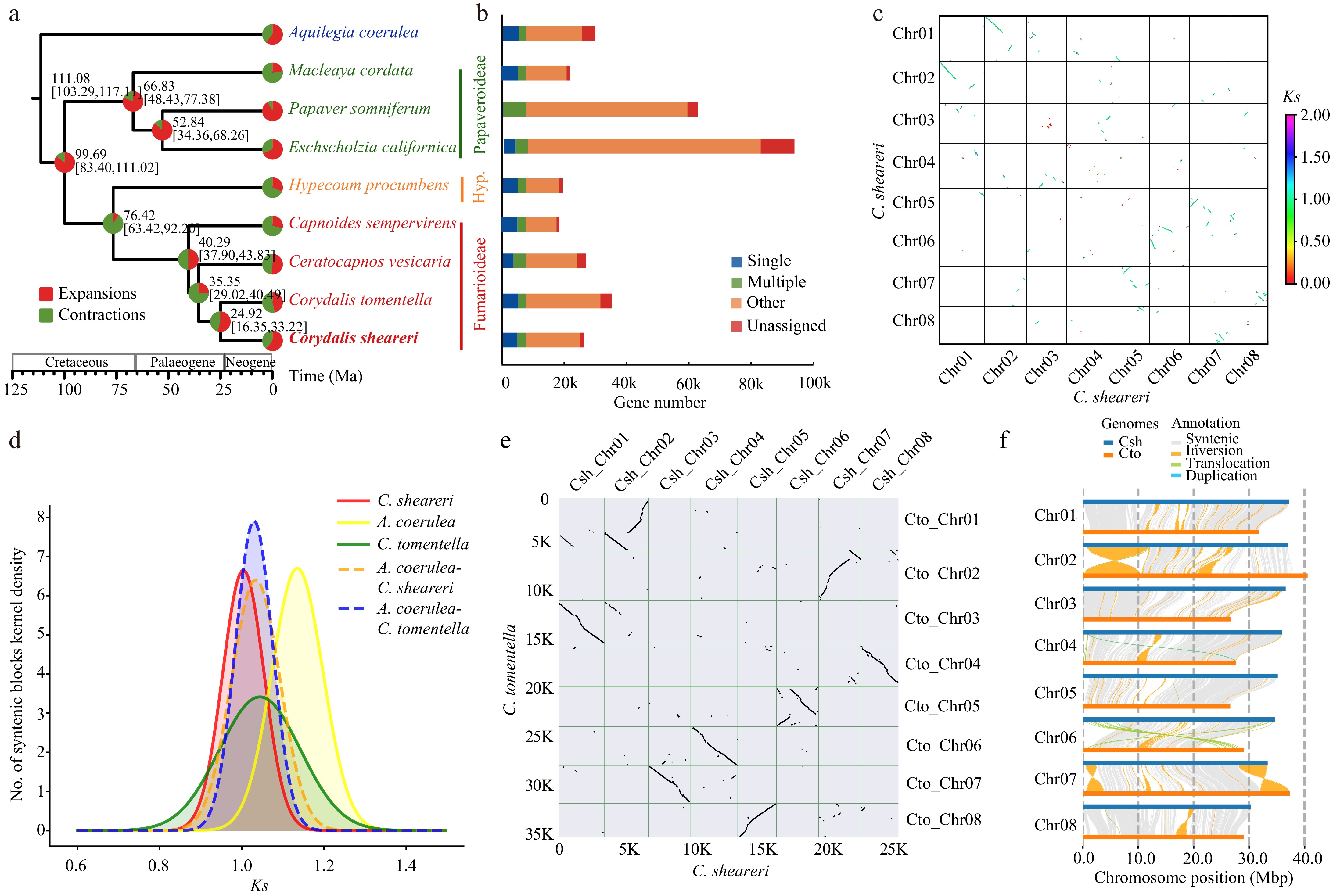
Figure 2.
Genome evolution of Corydalis sheareri. (a) Phylogenetic tree and divergence time estimation. Pal., Palaeogene; Neo., Neogene; Hyp., Hypecooideae. (b) Gene families identified in each species. (c) Collinearity within the genome of C. sheareri. (d) Ks distributions of anchor pairs for the paralogous genes of C. sheareri, C. tomentella and Aquilegia coerulea, and for orthologous genes between C. sheareri and C. tomentella, A. coerulea, respectively. (e) Collinearity analysis between C. sheareri and C. tomentella. (f) Structural variation detection between C. sheareri and C. tomentella performed by SyRI.
-
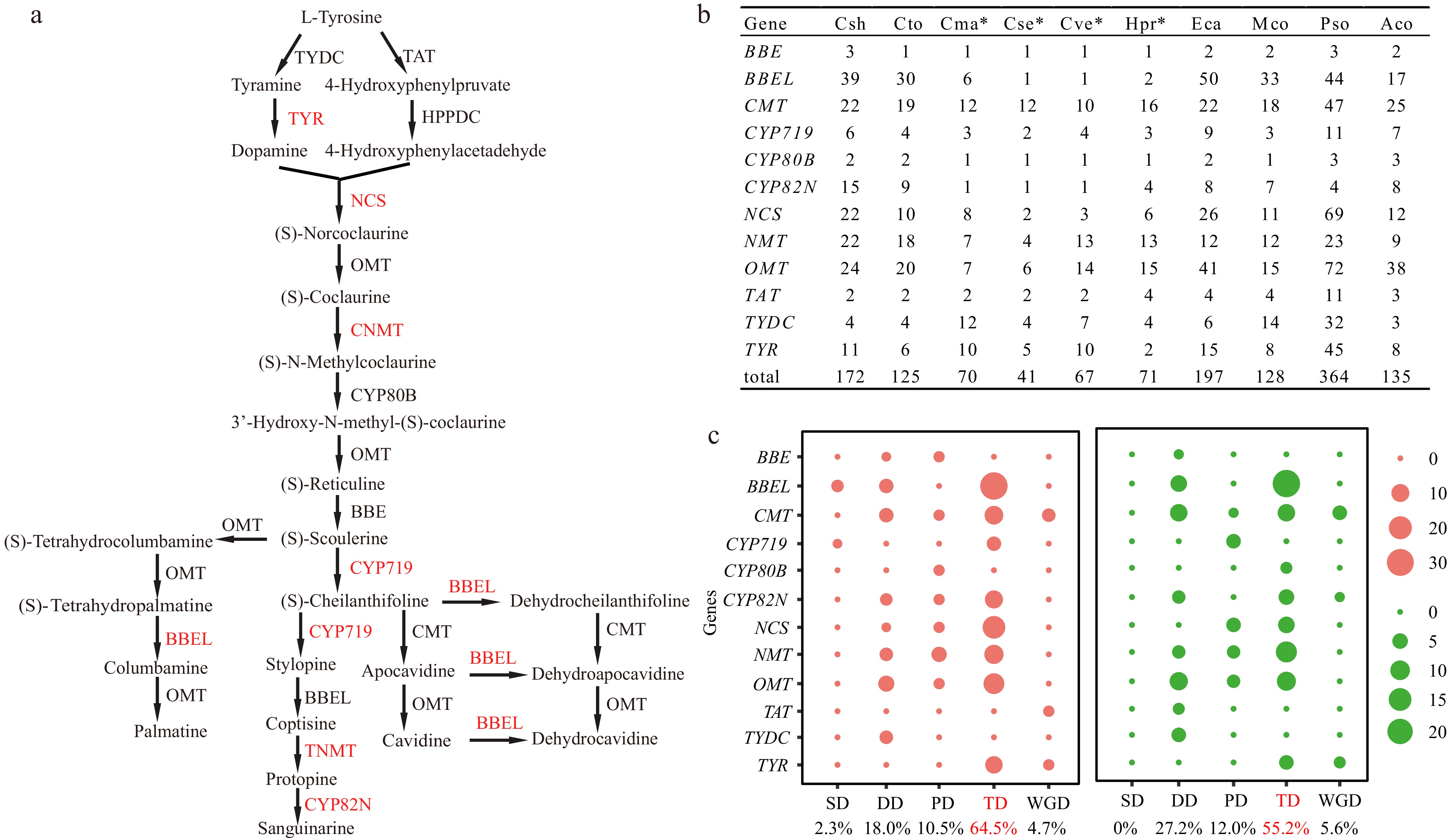
Figure 3.
BIAs biosynthesis related gene families identified in this study. (a) The biosynthetic pathway of benzylisoquinoline alkaloids (BIAs) in Corydalis. The species-specific tandem duplicated gene pairs or clusters in Corydalis identified in Fig. 5 and Table 2 were highlighted with red color. (b) The copy number of BIAs biosynthesis related gene families. Four species with transcriptome sequence are represented by asterisks. (c) Gene duplication type identified in C. sheareri (left) and C. tomentella (right). Abbreviations: BBE, berberine bridge enzyme; BBEL, berberine bridge enzyme-like; CMT, C-methyltransferase; CNMT, coclaurine N-methyltransferase; CYP, Cytochrome P450; HPPDC, 4-hydroxyphenylpyruvate decarboxylase; NCS, norcoclaurine synthase; NMT, N-methyltransferase; OMT, O-methyltransferase; TAT, tyrosine aminotransferase; TNMT, tetrahydroprotoberberine N-methyltransferase; TYDC, tyrosine decarboxylase; TYR, tyramine 3-hydroxylase. Aco, Aquilegia coerulea; Cma, Chelidonium majus; Cse, Capnoides sempervirens; Csh, Corydalis sheareri; Cto, C. tomentella; Cve, Ceratocapnos vesicaria; Eca, Eschscholzia californica; Hpr, Hypecoum procumbens; Mco, Macleaya cordata; Pso, Papaver somniferum. SD, Singleton; DD, dispersed duplications; PD, proximal duplications; TD, Tandem duplications; WGD, segmental/WGD duplications.
-
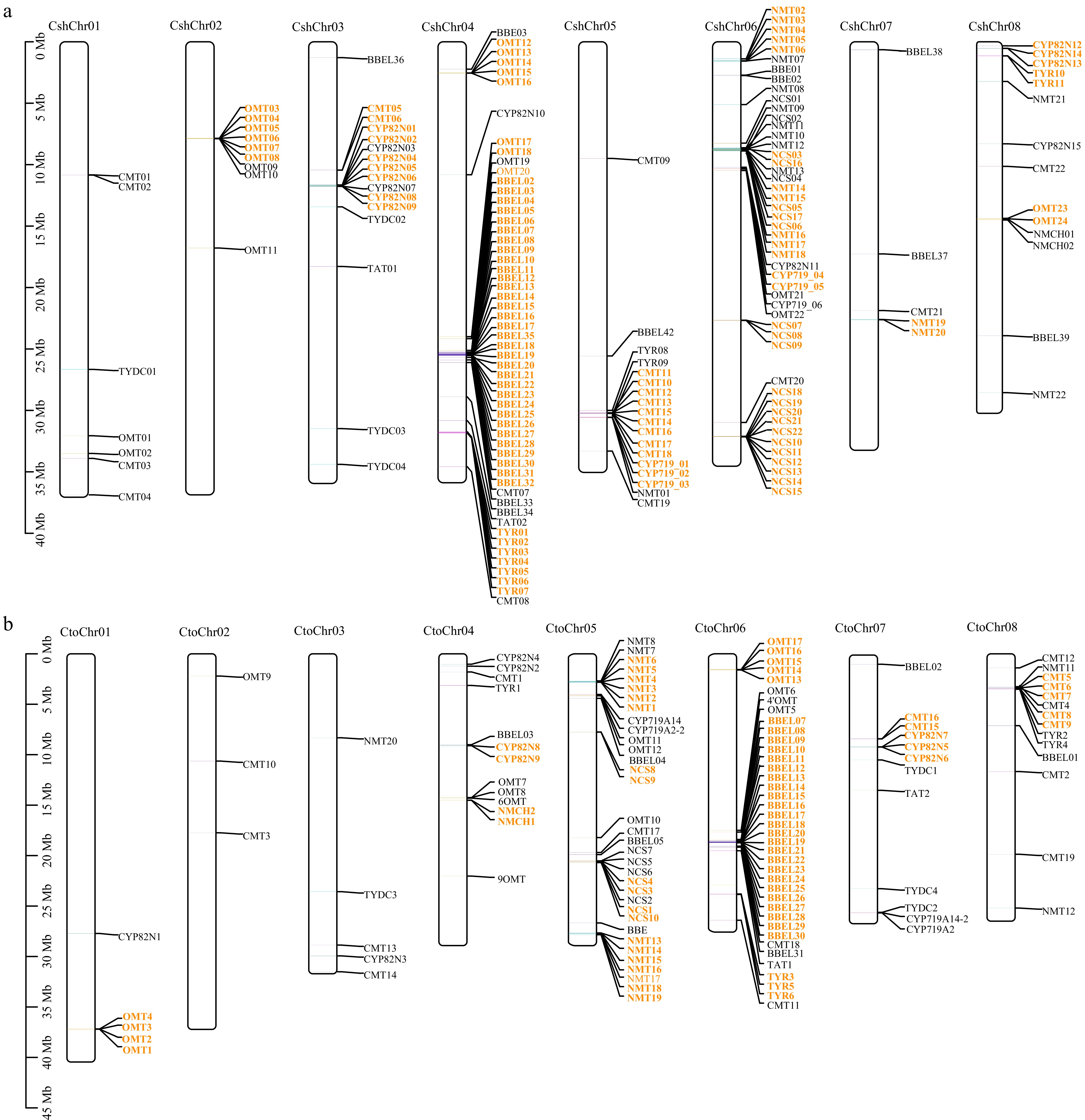
Figure 4.
Chromosome location and duplication event analyses of BIA biosynthesis genes in (a) C. sheareri and (b) C. tomentella. The chromosome number is indicated at the top of each chromosome. Tandem duplicated genes are indicated with orange color. The scale bar on the left indicates the length (mb) of chromosomes. The gene and species abbreviations were the same as Fig. 3. Chr, chromosome.
-
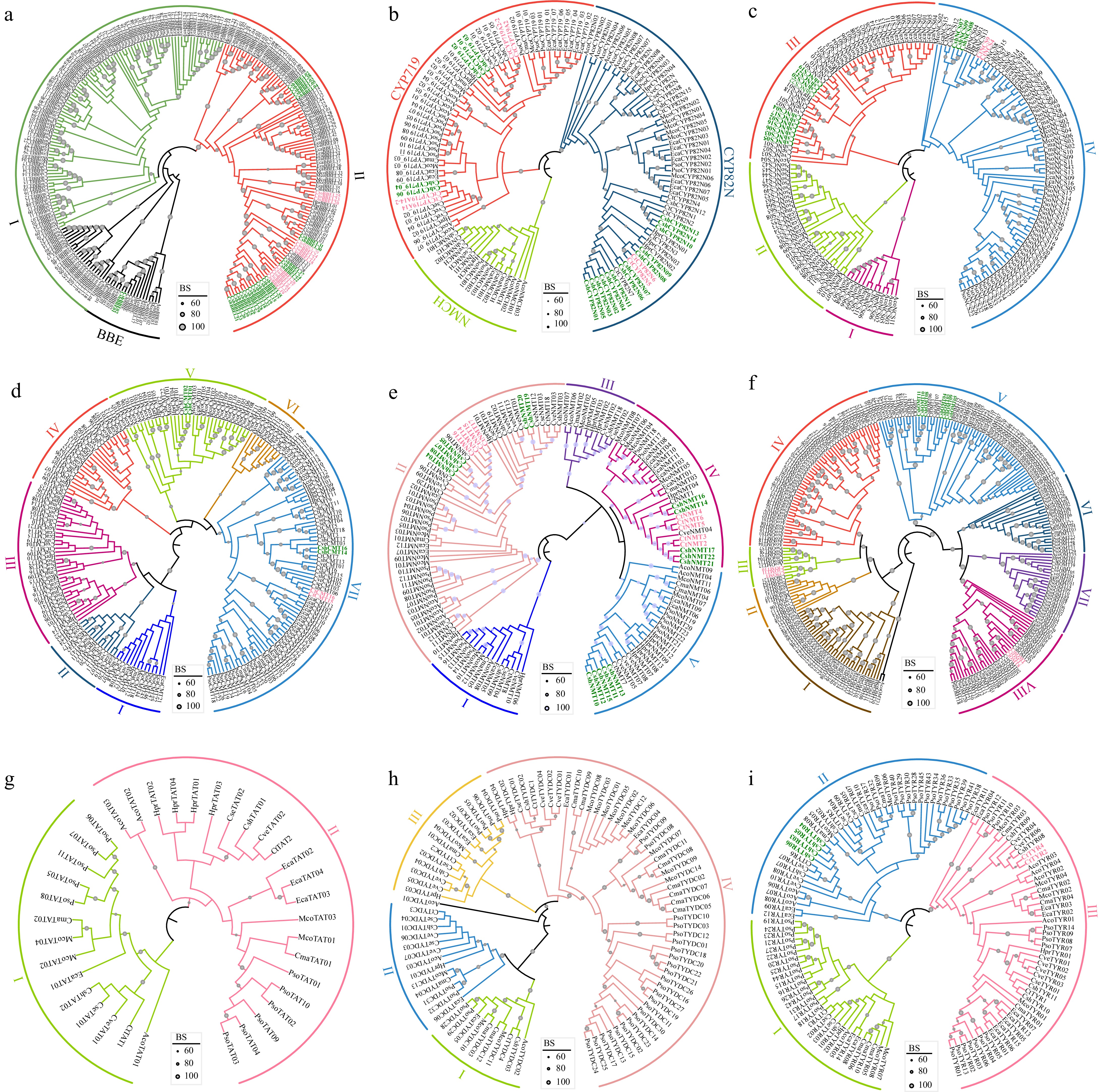
Figure 5.
Phylogenetic trees of BIAs biosynthesis genes. (a) BBE and BBEL, (b) CYP719, CYP80B (NMCH), and CYP82N, (c) NCS, (d) CMT, (e) NMT, (f) OMT, (g) TAT, (h) TYDC, and (i) TYR. Corydalis sheareri and C. tomentella – specific monophyletic groups are highlighted with green and pink color, respectively. The gene and species abbreviations were the same as Fig. 3.
-
Analytical process Characteristic C. sheareri Genome survey Genome size (flow cytometry) (Mb) 580 Genome size (k-mer spectrum) (Mb) 261 Assembly_csh v1.0 Number of contigs 2,713 Assembly size (Mb) 550 Contig N50 (Mb) 9.18 Shortest contig (bp) 16,037 Largest contig (bp) 25,090,490 Assembly_csh v2.0 Total number of contigs 179 Assembly size (Mb) 282 Contig N50 (Mb) 11.39 Number of pseudochromosomes 8 GC content 37.11% Annotation Number of protein-coding genes 26,287 Mean gene length (bp) 5,092 Mean CDS length (bp) 1,440 Complete BUSCOs (C) 1,567 Percentage of repeat sequences (%) 44.47 Table 1.
Genome assembly and annotation of Corydalis sheareri.
-
Gene pairs or clusters Ka Ks Ka/Ks Similarity CshBBEL03, CshBBEL04, CshBBEL05 0.1656 0.4321 0.38315 97.54% CshBBEL30, CshBBEL31 0.1090 0.3349 0.32543 98.73% CshCYP719_01, CshCYP719_02,
CshCYP719_030.0841 0.3688 0.22803 98.81% CshNCS07, CshNCS08, CshNCS09 0.0546 0.2236 0.24430 99.11% CshNCS19, CshNCS20, CshNCS21 0.1325 0.8527 0.15543 94.61% CshNMT19, CshNMT20 0.1435 0.4799 0.29900 98.68% CshTYR03, CshTYR04, CshTYR05, CshTYR06 0.1635 0.373 0.43823 97.79% CtCYP719A2, CtCYP719A2-2 0.0841 0.3808 0.22096 97.73% CtCYP82N5, CtCYP82N6 0.0913 0.2672 0.34165 97.79% Table 2.
Selective pressure and sequence similarity of the species-specific tandem duplicated gene pairs or clusters in Corydalis.
Figures
(5)
Tables
(2)