-
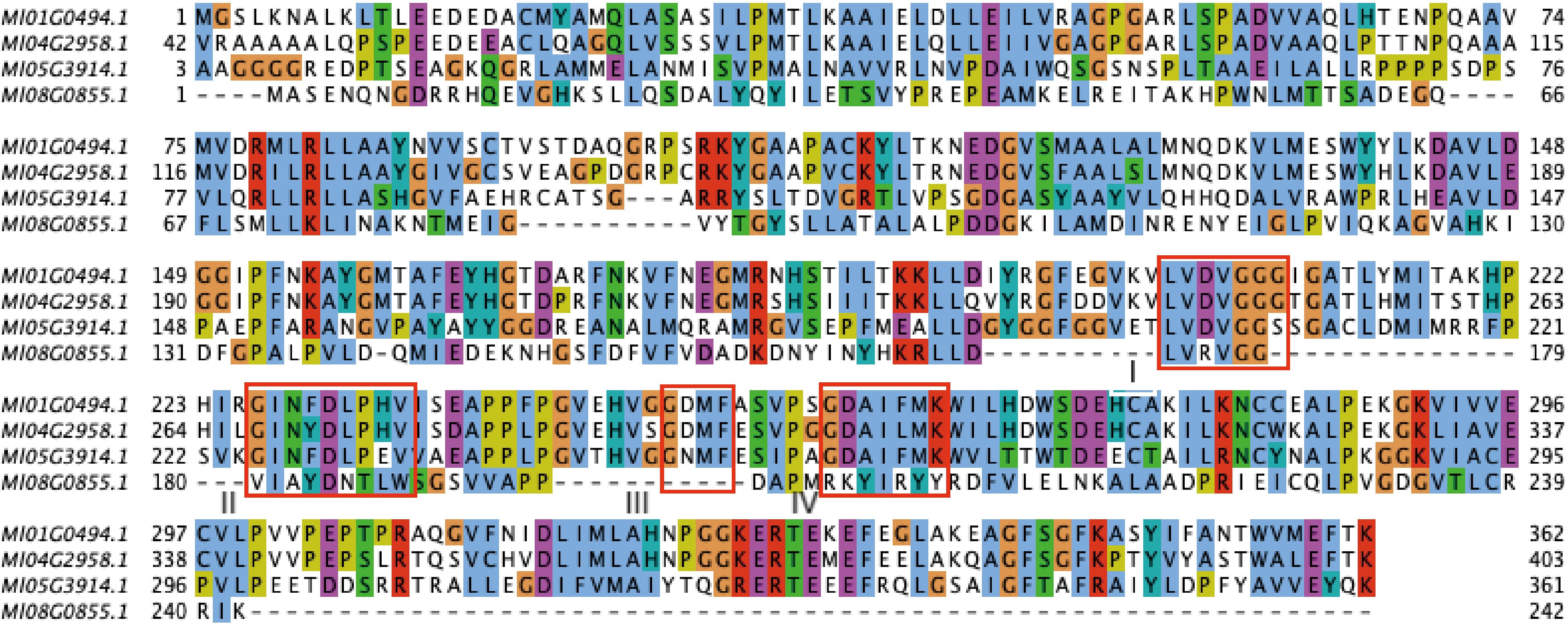
Figure 1.
Multiple sequence alignment of MlOMTs and AtCOMT1. Asterisks indicate conserved residues, namely substrate binding sites (M128, V314, I317, M318), SAM binding sites (V215, D229, G248, K263), a critical residue (E327) in AtOMT1. Conserved motifs I–IV are underlined. At, Arabidopsis thaliana.
-
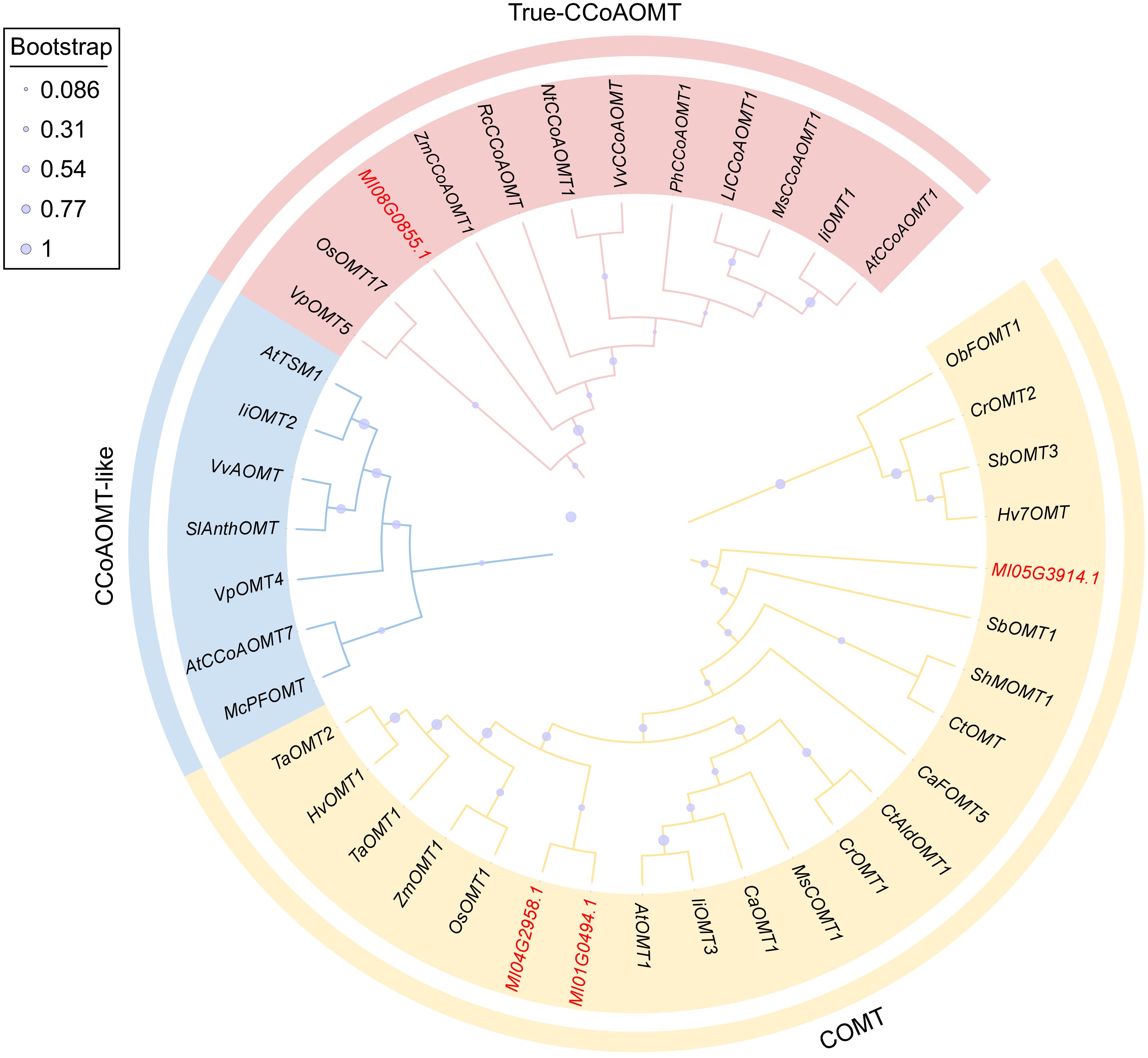
Figure 2.
Phylogenetic analysis of MlOMTs and other plant OMT proteins. The GenBank accession numbers of OMT proteins from other plants are listed in Supplementary Table S2.
-
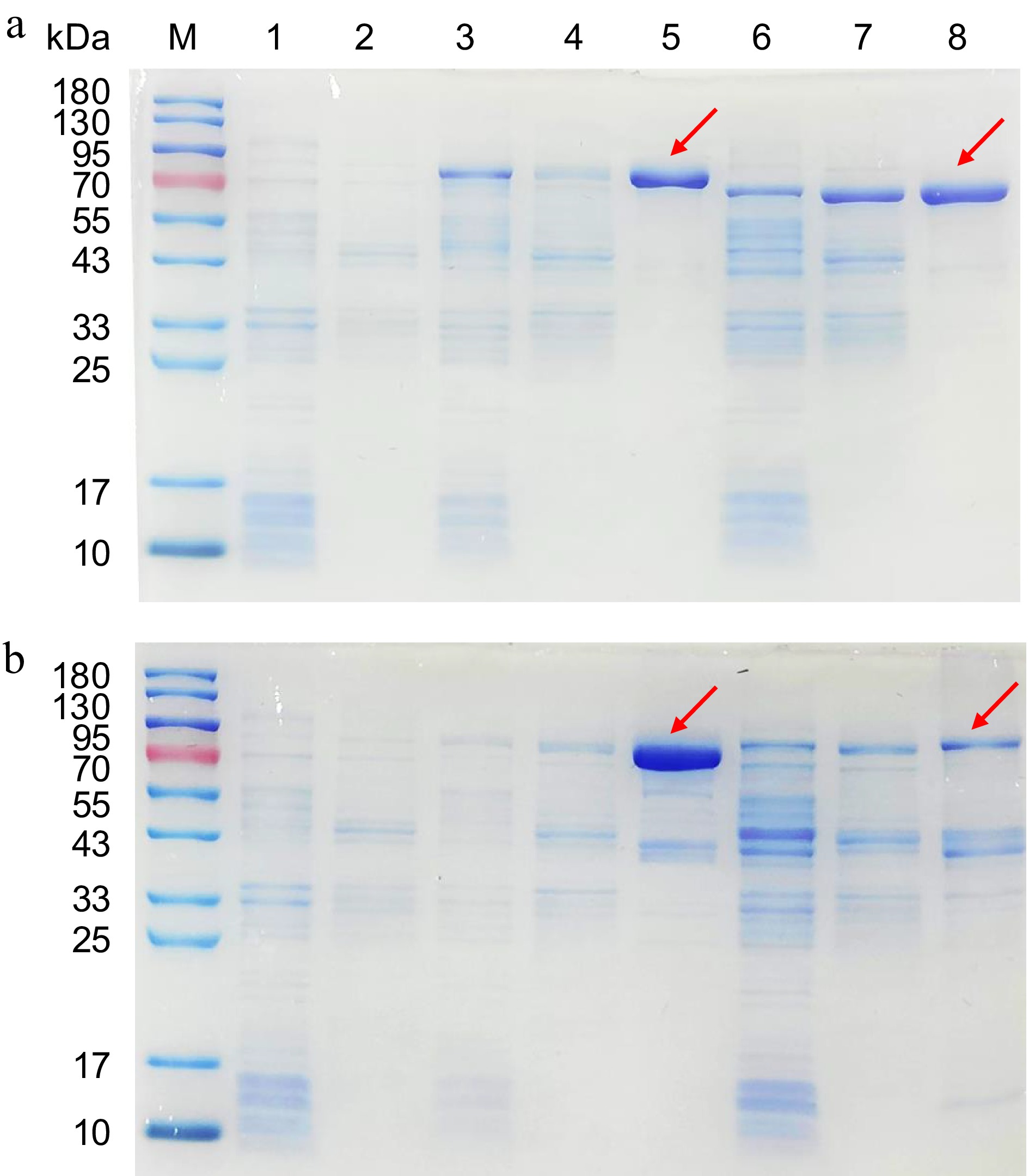
Figure 3.
SDS-PAGE analysis of recombinant Ml01G0494, Ml08G0855, Ml04G2958, and Ml05G3914 proteins. (a) M: protein molecular marker; Lane 1: pMAL-c4x sediment (MBP ≈ 42 kDa); Lane 2: pMAL-c4x supernatant; Lane 3: Ml01G0494 sediment; Lane 4: Ml01G0494 supernatant; Lane 5: Ml01G0494 purified recombinant protein; Lane 6: Ml08G0855 sediment; Lane 7: Ml08G0855 supernatant; Lane 8: Ml08G0855 purified recombinant protein. (b) M: protein molecular marker; Lane 1: pMAL-c4x sediment; Lane 2: pMAL-c4x supernatant; Lane 3: Ml04G2958 sediment; Lane 4: Ml04G2958 supernatant; Lane 5: Ml04G2958 purified recombinant protein; Lane 6: Ml05G3914 sediment; Lane 7: Ml05G3914 supernatant; Lane 8: Ml05G3914 purified recombinant protein.
-
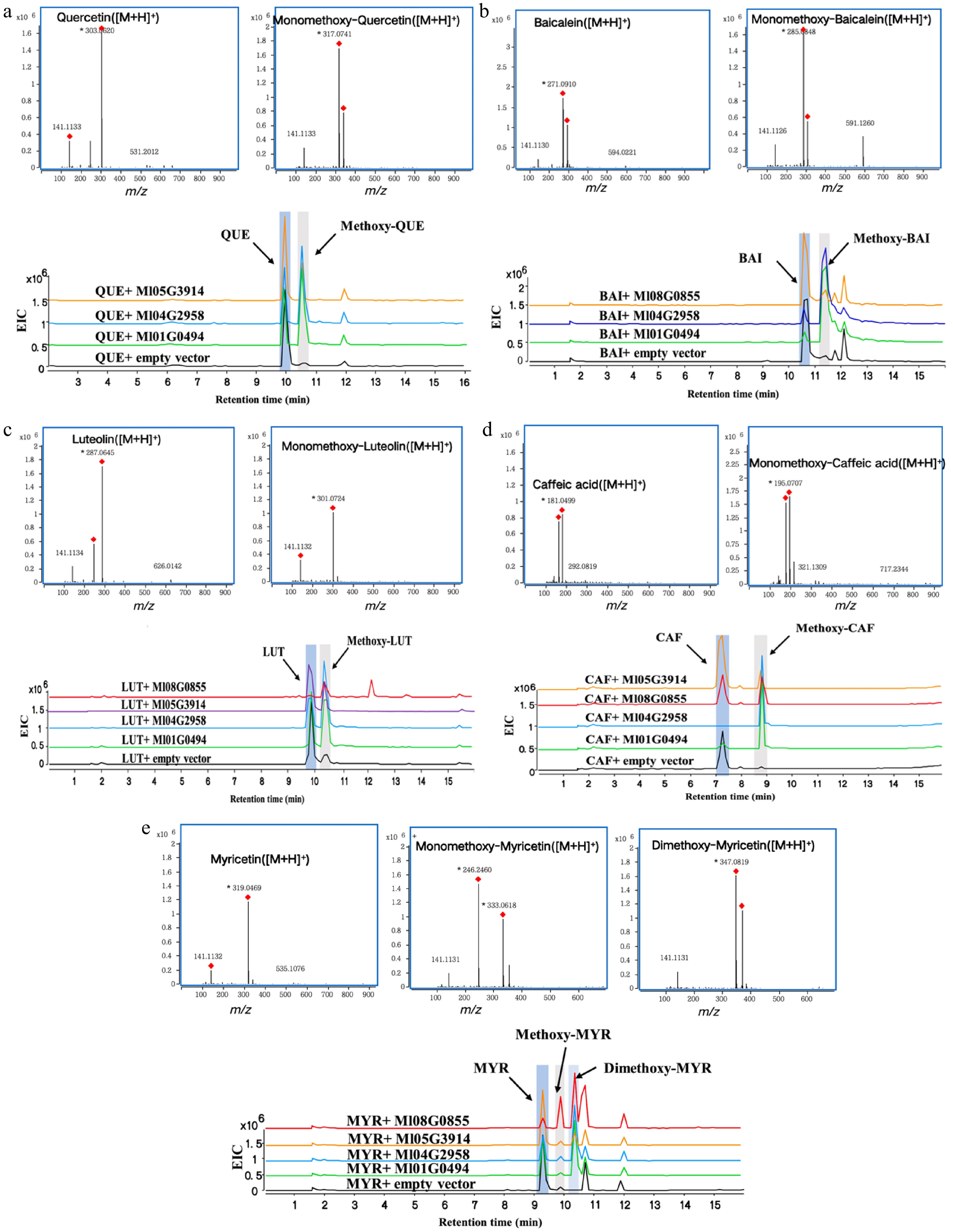
Figure 4.
HPLC-Q TOF-MS/MS analysis of enzymatic reactions using quercetin, baicalin, luteolin, caffeic acid, and myricetin as substrates respectively. (a) Quercetin (QUE), (b) baicalin (BAI), (c) luteolin (LUT), (d) caffeic acid (CAF), (e) myricetin (MYR).
-
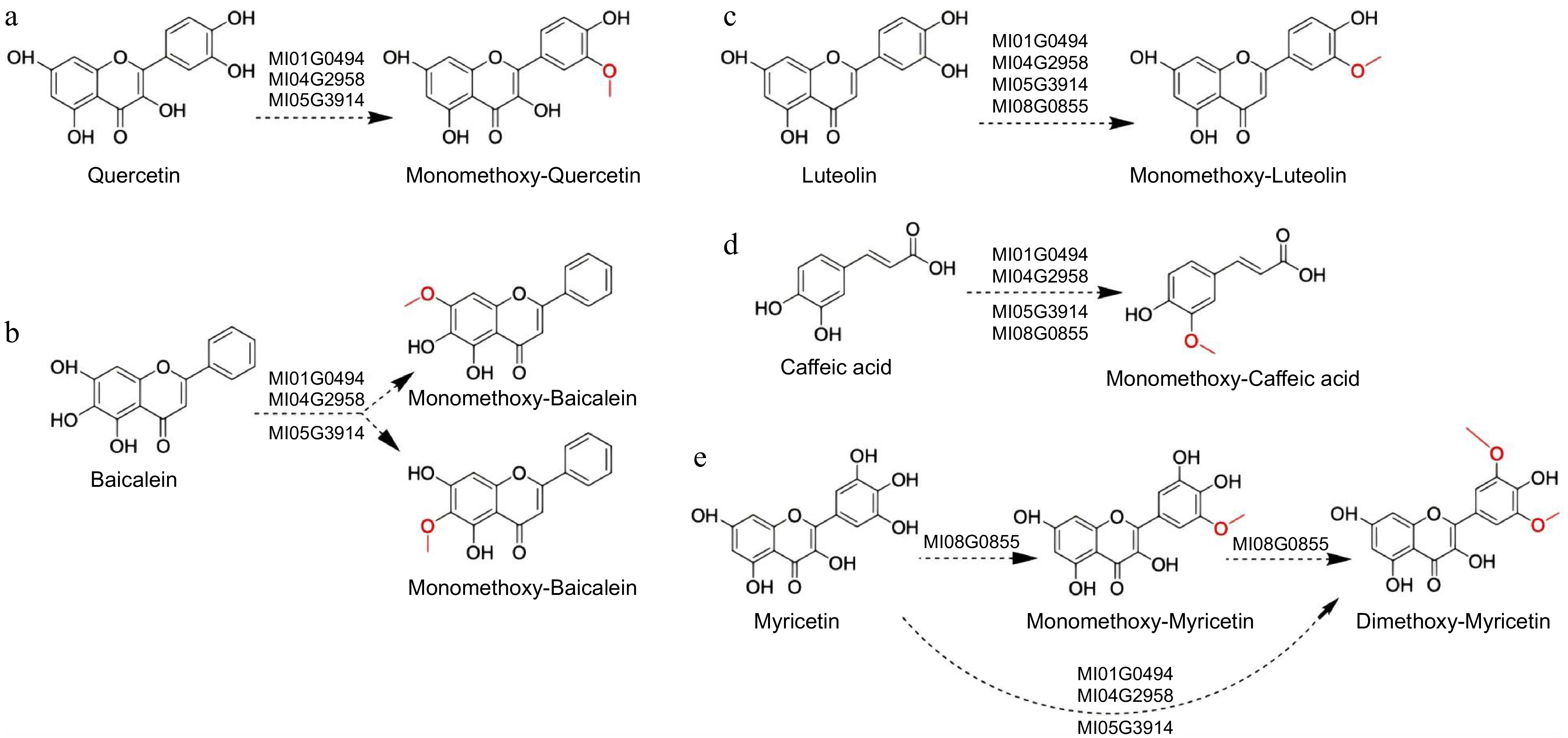
Figure 5.
Results of the Ml01G0494, Ml04G2958, Ml08G0855, and Ml05G3914 assays with flavonoid and caffeic acid. The dashed arrows indicate the possible catalytic sites based on the literature. (a) Quercetin, (b) baicalin, (c) luteolin, (d) caffeic acid, (e) myricetin.
Figures
(5)
Tables
(0)