-
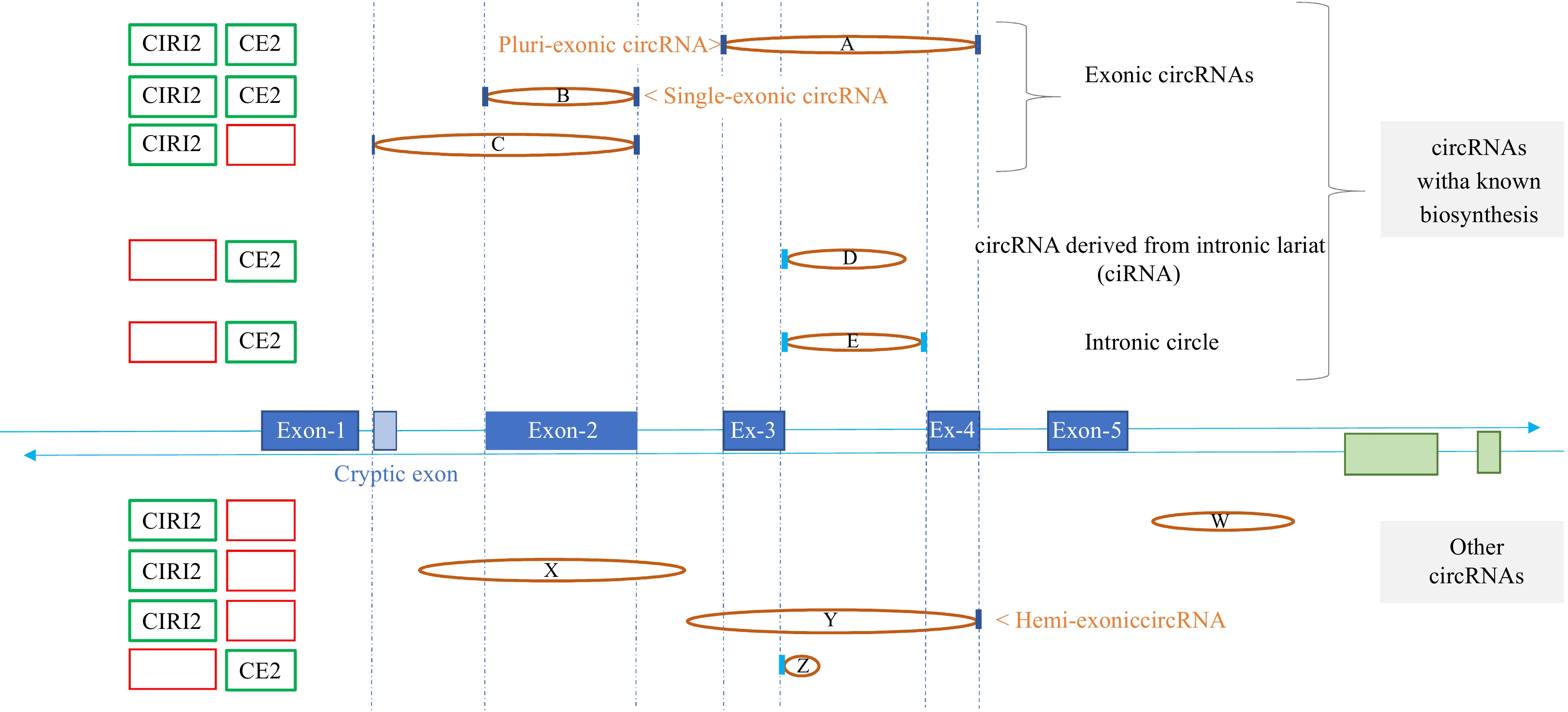
Figure 1.
Genomic region exemplifying variability in circRNA detection across computational tools. This figure illustrates a genomic region where reads corresponding to nine circular junctions were detected using several circRNA tools. The central section displays the genomic structure, featuring one gene (in blue) with five exons and a cryptic exon, as well as the terminal exons of a gene (in green) located on the reverse strand. Vertical dashed blue lines mark the 5'-Start and 3'-End of exons concerned by circRNAs. Circular junctions, identified by paired coordinates within the reads, are represented as ellipses connecting the two points. If a coordinate aligns with the 5'-Start or the 3'-End of an exon, it is marked with a dark blue vertical line; if it aligns with an intron boundary, it is marked in cyan blue. The five circRNAs above the genomic structure (circRNAs A to E) are associated with well-established biosynthetic pathways, while the four circRNAs below (circRNAs W to Z) lack classification within known categories, such as exonic circRNAs involving annotated exons (circRNAs A to C), intronic lariat-derived circRNA (circRNA-D), or intronic circle (circRNA-E). CIRI2 successfully identifies six of these nine circRNAs, while CIRCexplorer2 (CE2) detects only five. The presence or absence of each circRNA in the outputs of CE2 and CIRI2 is visually represented on the left by green and red boxes, respectively. Finally, it is important to emphasize that the full sequence of a circRNA can only be inferred based on its circular junction for certain categories, such as ciRNA (circRNA-D), intronic circle (circRNA-E), and single-exonic circRNA (circRNA-B).
-
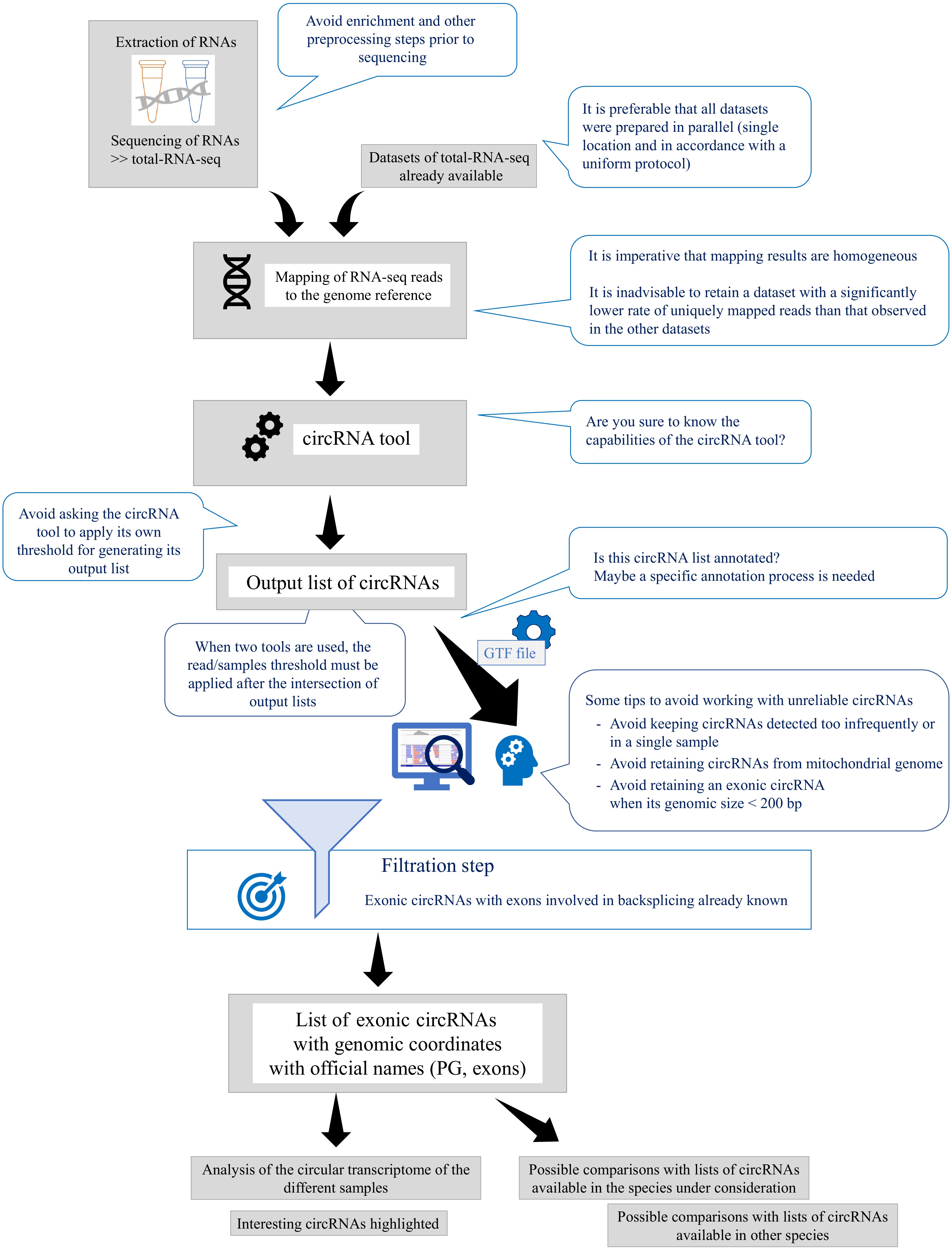
Figure 2.
Recommendations for enhancing user understanding of circRNAs, tools, and output lists. This figure provides practical recommendations to assist users in developing a more nuanced understanding of circRNAs, the tools used to detect them, and the interpretation of output lists. Emphasis is placed on the utility of GTF files for annotating circRNAs, as these files include essential exon coordinates. The term 'PG' refers to parental genes, highlighting their role in circRNA analysis.
Figures
(2)
Tables
(0)