-
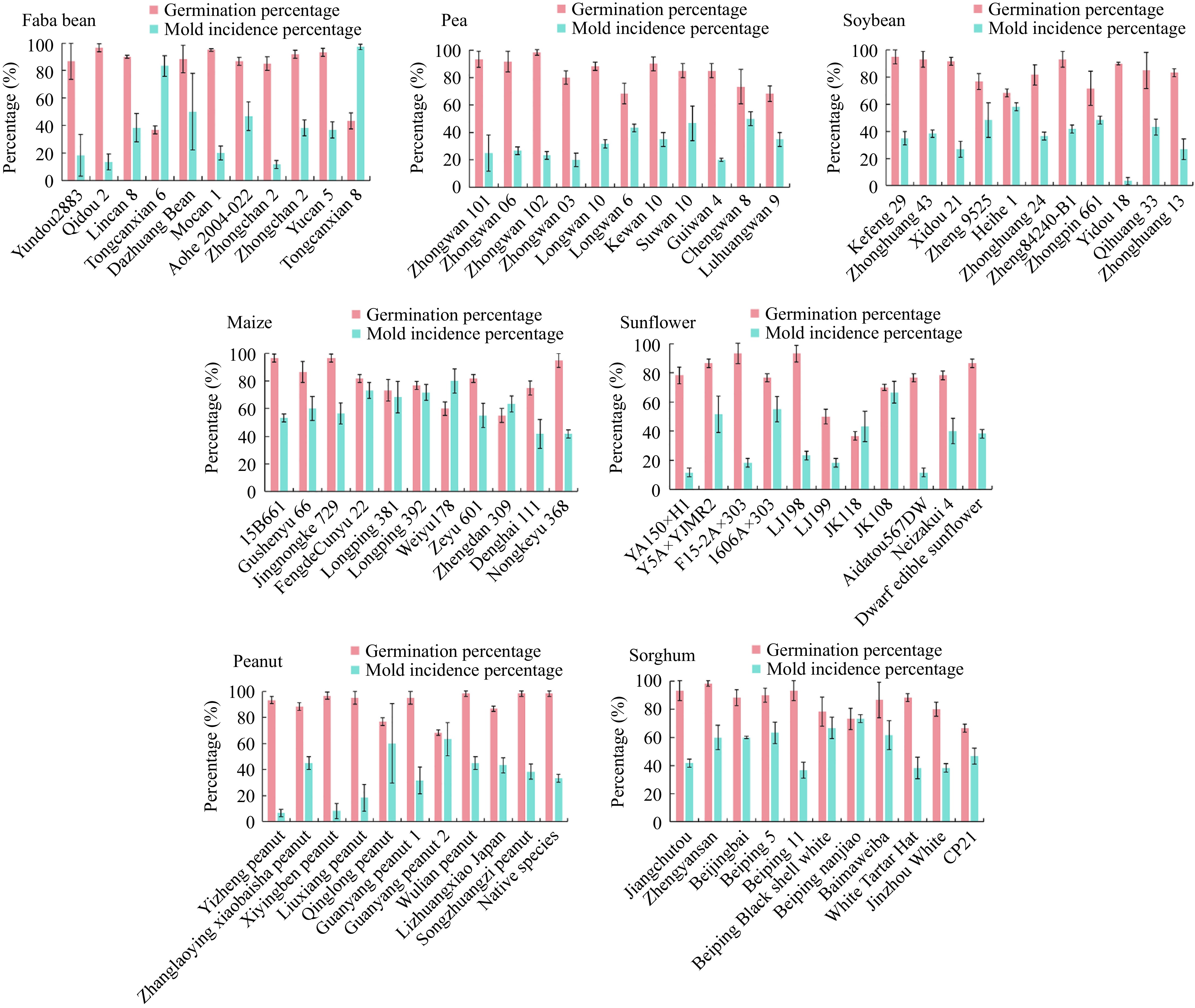
Figure 1.
Detection germination percentage and mold incidence percentage in seeds from 11 varieties of each faba bean, pea, soybean, maize, sunflower, peanut, and sorghum, germinated according to the ISTA protocol. Values are presented as means ± SD (n = 3).
-
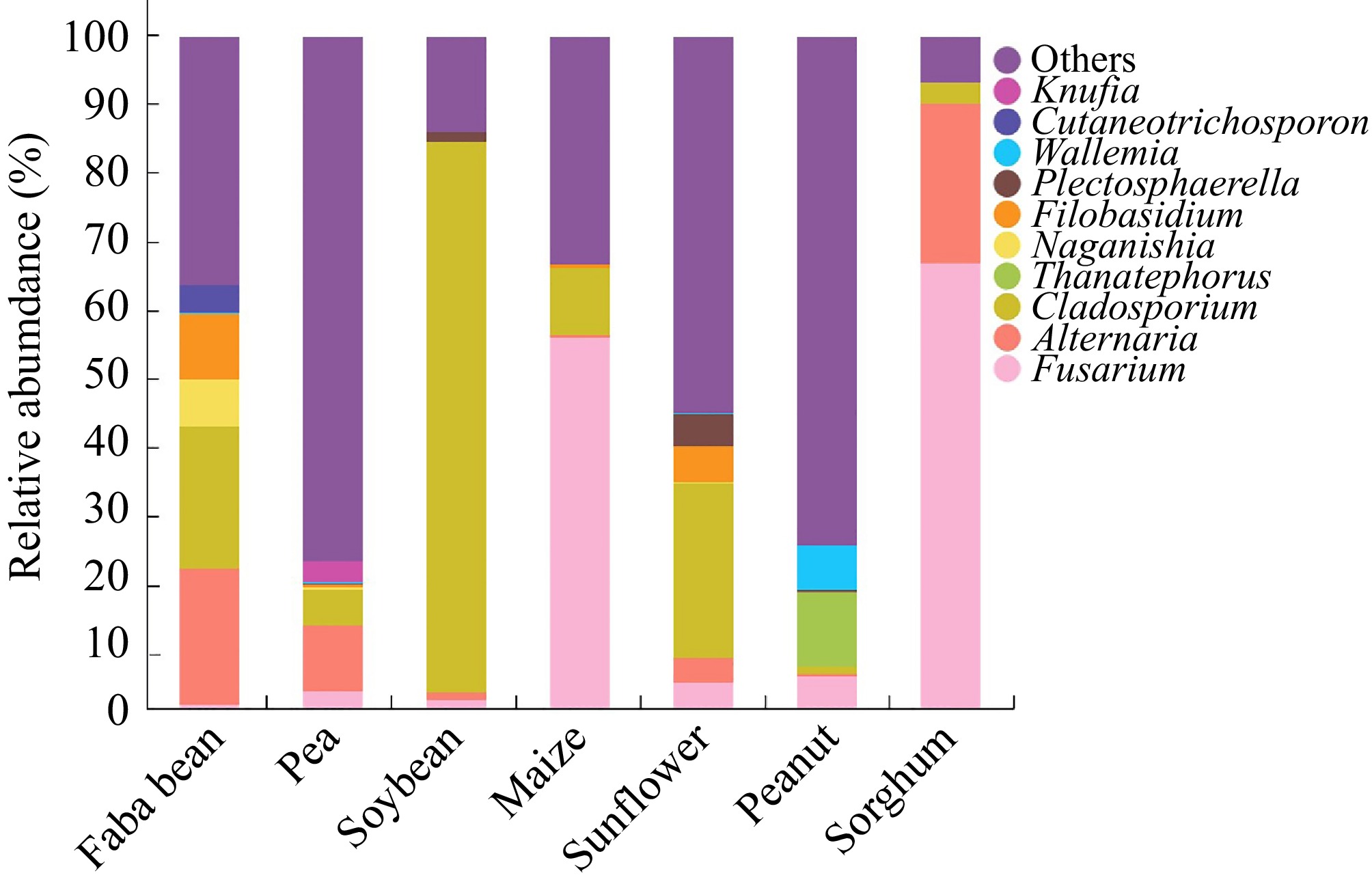
Figure 2.
Identification and classification of fungi associated with seeds of faba bean ('Tongcanxian 6'), pea ('Zhongwan 06'), soybean ('Zhonghuang 13'), maize ('Nongkeyu 368'), sunflower ('Dwarf edible sunflower'), peanut ('Native species'), and sorghum ('CP21') as determined by high-throughput sequencing.
-
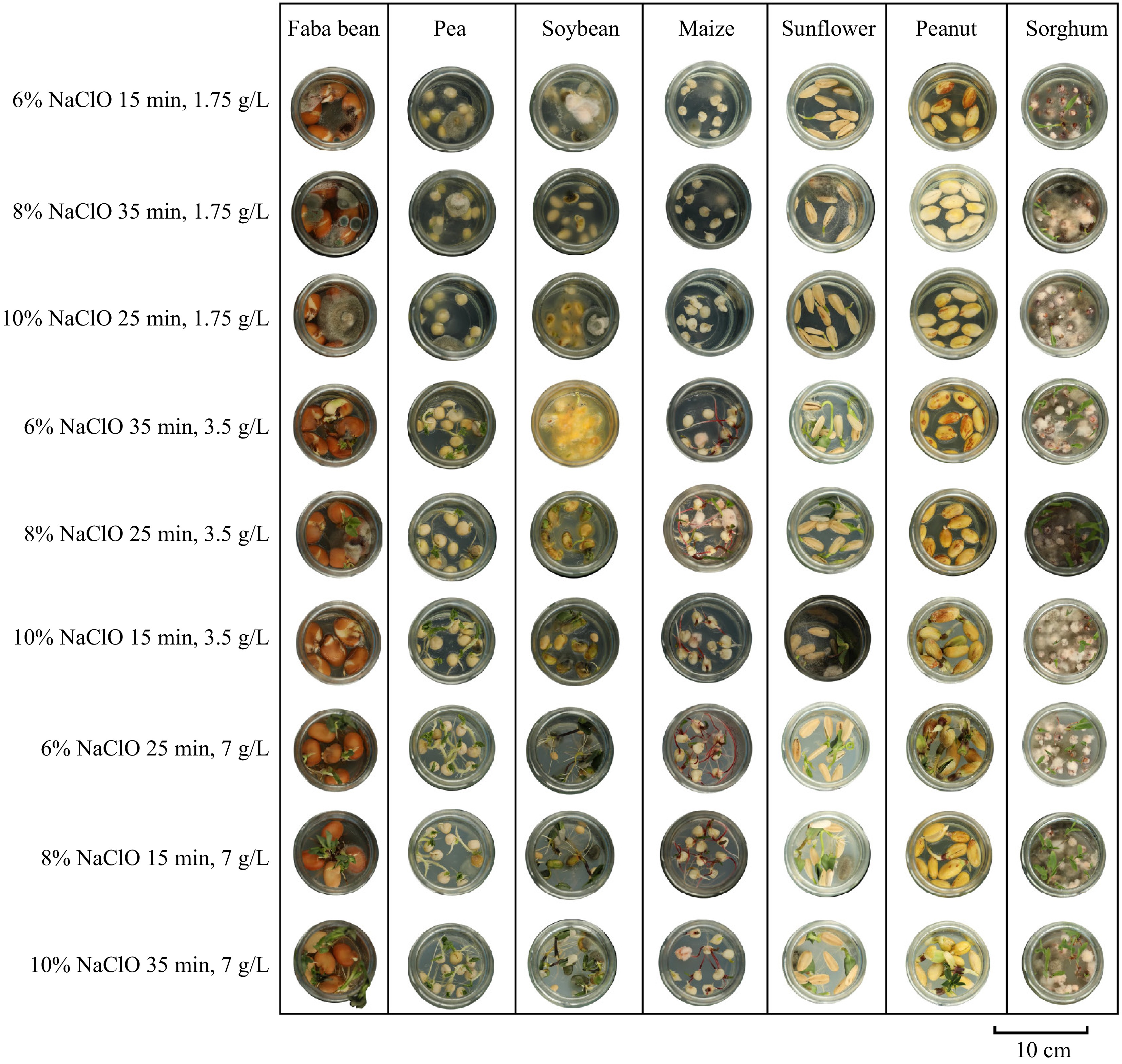
Figure 3.
Assessment of fungal contamination on the germination of faba bean ('Tongcanxian 6'), pea ('Zhongwan 06'), soybean ('Zhonghuang 13'), maize ('Nongkeyu 368'), sunflower ('Dwarf edible sunflower'), peanut ('Native species'), and sorghum ('CP21') seeds treated with NaClO solutions at concentrations of 6%, 8%, and 10%, disinfection times of 15 min, 25 min, and 35 min, and agar concentrations of 1.75 g/L, 3.5 g/L, and 7 g/L in the culture medium, with a pH range of 5.8–6.0.
-
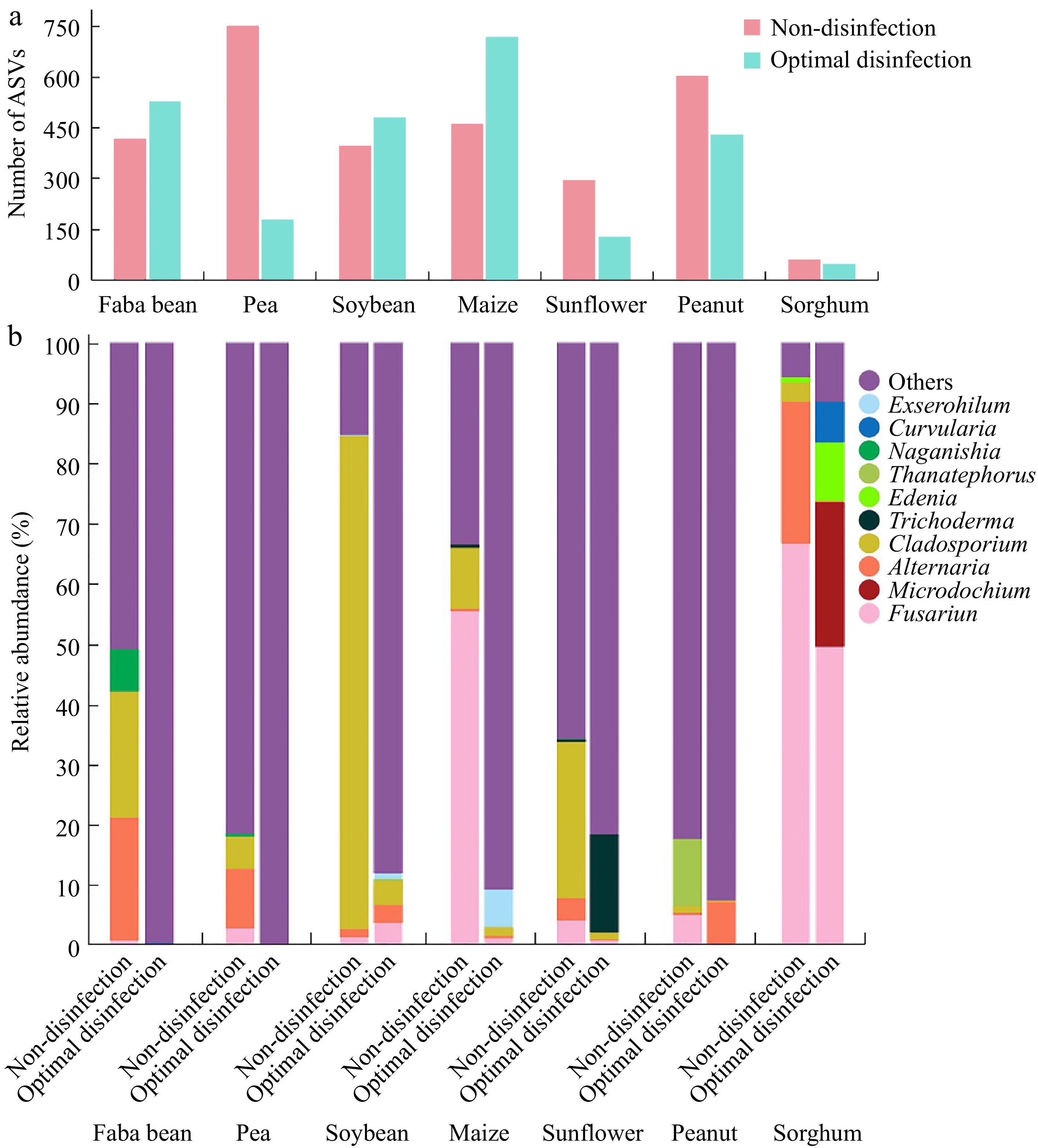
Figure 4.
Composition of the major fungi in faba bean ('Tongcanxian 6'), pea ('Zhongwan 06'), soybean ('Zhonghuang 13'), maize ('Nongkeyu 368'), sunflower ('Dwarf edible sunflower'), peanut ('Native species'), and sorghum ('CP21') seeds treated with optimal disinfection protocol and without disinfection as determined through high-throughput sequencing. (a) The number of Amplicon Sequence Variants (ASVs). (b) Relative abundance of major fungal genera.
-
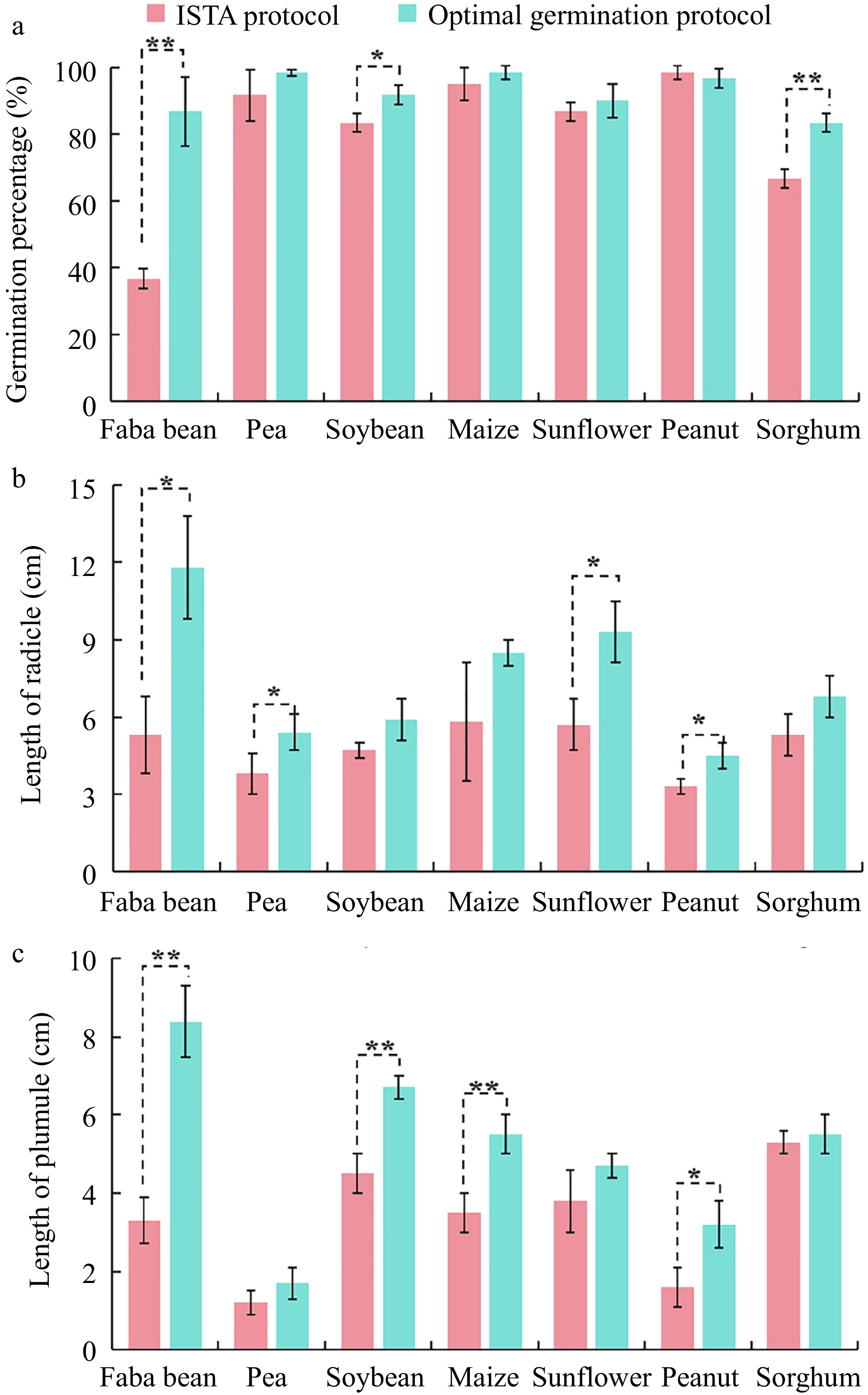
Figure 5.
Comparison of the effect of the optimal disinfection protocol and the ISTA protocol on (a) germination percentage, (b) radicle length, and (c) plumule length of faba bean ('Tongcanxian 6'), pea ('Zhongwan 06'), soybean ('Zhonghuang 13'), maize ('Nongkeyu 368'), sunflower ('Dwarf edible sunflower'), peanut ('Native species'), and sorghum ('CP21') seeds. Values are presented as means ± SD (n = 3). Significant differences were determined using one-way ANOVA (*, p < 0.05; **, p < 0.01).
-
Faba bean Pea Soybean Maize Sunflower Peanut Sorghum Cladosporium Alternaria Cladosporium Fusarium Cladosporium Thanatephorus Fusarium Alternaria Cladosporium Humicola Cladosporium Filobasidium Wallemia Alternaria Filobasidium Didymella Fusarium Sarocladium Plectosphaerella Fusarium Cladosporium Naganishia Knufia Plectosphaerella Filobasidium Fusarium Nigrospora Ramichloridium Cutaneotrichosporon Fusarium Hannaella Trichoderma Alternaria Cladosporium Edenia Talaromyces Vacuiphoma Alternaria Alternaria Golovinomyces Neocosmospora Ophiosphaerella Vishniacozyma Botrytis Aspergillus Papiliotrema Botrytis Paramyrothecium Phaeosphaeria Aspergillus Celosporium Rhodosporidiobolus Rhizopus Penicillium Talaromyces Hannaella Fusarium Nigrospora Acremonium Aspergillus Rhizopus Malassezia Moesziomyces Didymella Dactylonectria Malassezia Vishniacozyma Hanseniaspora Plectosphaerella Bipolaris Table 1.
The fungal genera of the top 10 most abundant detected in seeds of faba bean ('Tongcanxian 6'), pea ('Zhongwan 06'), soybean ('Zhonghuang 13'), maize ('Nongkeyu 368'), sunflower ('Dwarf edible sunflower'), peanut ('Native species'), and sorghum ('CP21') as identified through high-throughput sequencing.
-
Fungi Organism Sequencing Crop species Faba bean Pea Soybean Maize Sunflower Peanut Sorghum Alternaria Seedlings Sanger + Tissue blocks Sanger + + + Seeds High-throughput + + + + + + Cladosporium Seedlings Sanger + + Tissue blocks Sanger + + Seeds High-throughput + + + + + + + Fusarium Seedlings Sanger + + Tissue blocks Sanger + + + Seeds High-throughput + + + + + + + + represents the genus of the fungi detected. Table 2.
The occurrence of Alternaria, Cladosporium, and Fusarium identified in seedlings and tissue blocks through Sanger sequencing, and in entire seeds through high-throughput sequencing of faba bean ('Tongcanxian 6'), pea ('Zhongwan 06'), soybean ('Zhonghuang 13'), maize ('Nongkeyu 368'), sunflower ('Dwarf edible sunflower'), peanut ('Native species'), and sorghum ('CP21').
-
Specie NaClO concentration (%) Disinfection time (min) Agar concentration (g/L) Faba bean 10 35 7 Pea 10 35 7 Soybean 8 15 7 Maize 6 25 7 Sunflower 10 35 7 Peanut 6 25 7 Sorghum 10 35 7 Table 3.
The optimal germination protocol for faba bean ('Tongcanxian 6'), pea ('Zhongwan 06'), soybean ('Zhonghuang 13'), maize ('Nongkeyu 368'), sunflower ('Dwarf edible sunflower'), peanut ('Native species'), and sorghum ('CP21'), including NaClO solution concentrations, disinfection times, and agar concentrations in the culture medium.
Figures
(5)
Tables
(3)