-
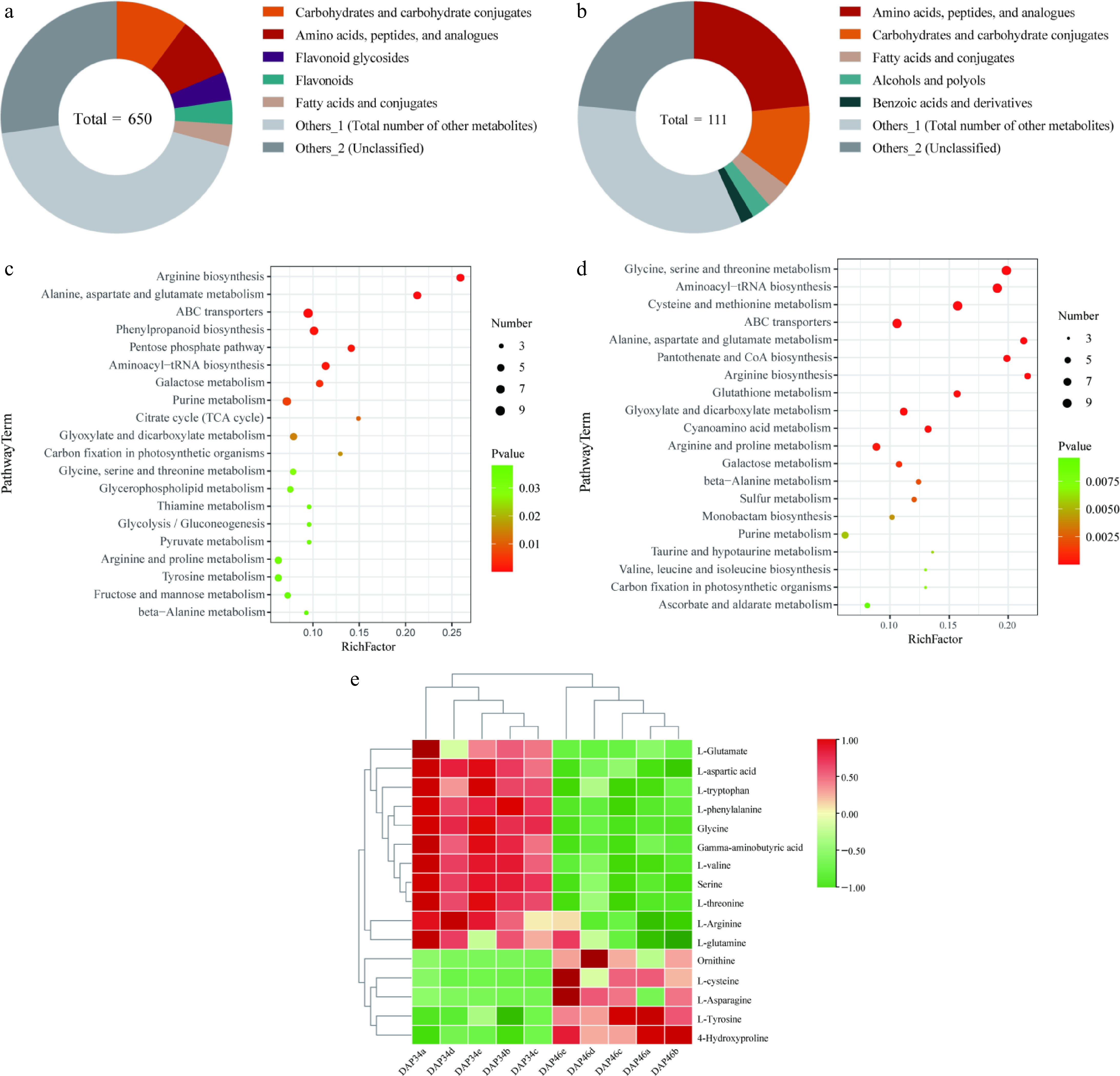
Figure 1.
Extensive targeted metabolomics analysis revealing the changes in amino acid contents before and after the quality transformation of Wucai. Classification of subclasses of differentially accumulated metabolites (DAMs) in (a) liquid chromatography-tandem mass spectrometry (LC-MS/MS), and (b) gas chromatography-tandem mass spectrometry (GC-MS/MS). Enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways associated with DAMs in (c) LC-MS/MS, and (d) GC-MS/MS. (e) Heat map showing the abundance of flavor amino acids detected in the metabolome.
-
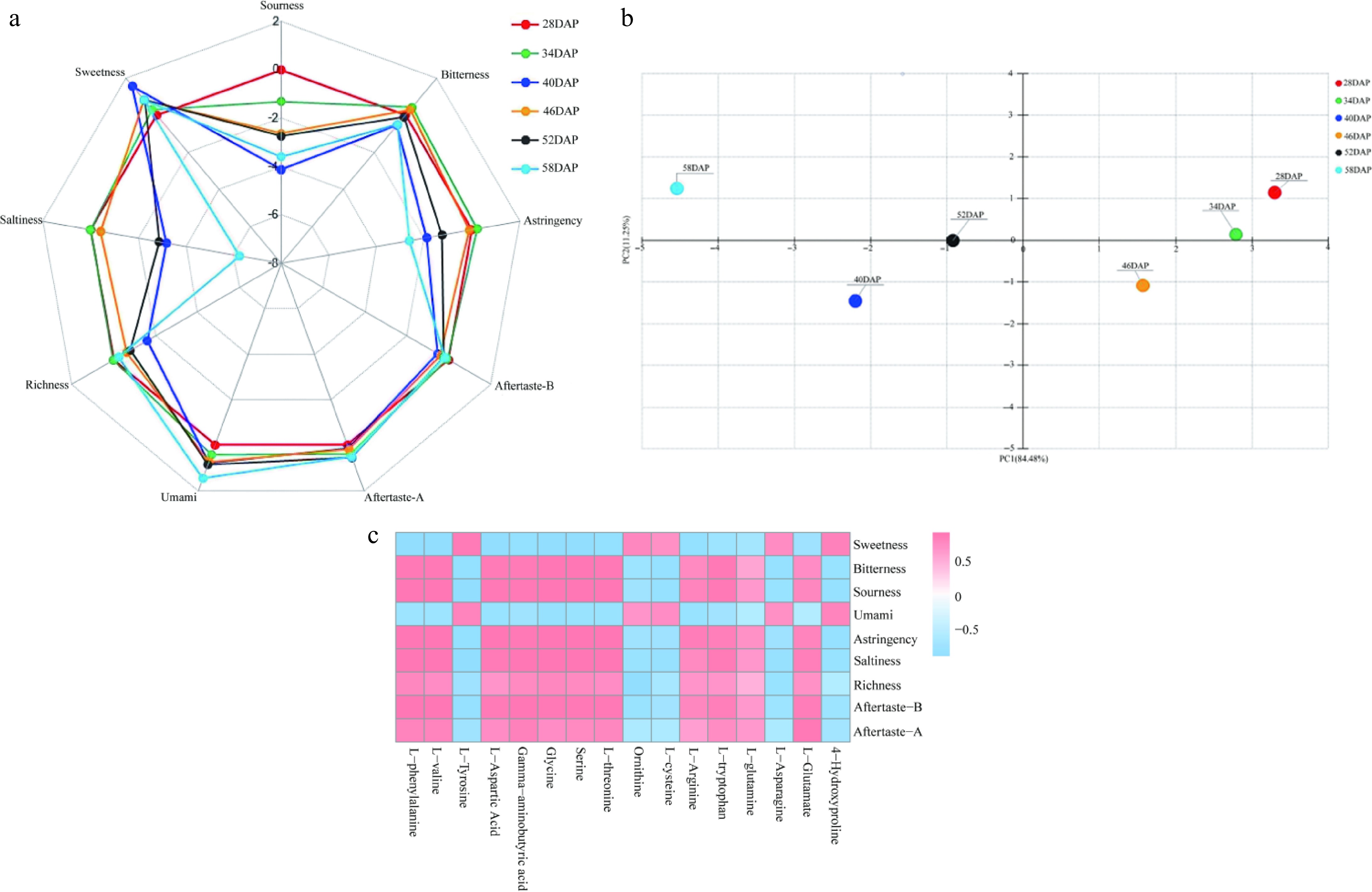
Figure 2.
Changes in taste attributes before and after the quality transformation of Wucai as determined by electronic tongue analysis. (a) Radar plot of electronic tongue analysis results. (b) Principal component analysis plot of electronic tongue analysis results. (c) Heat map showing the correlation between electronic tongue response values and flavor amino acids.
-
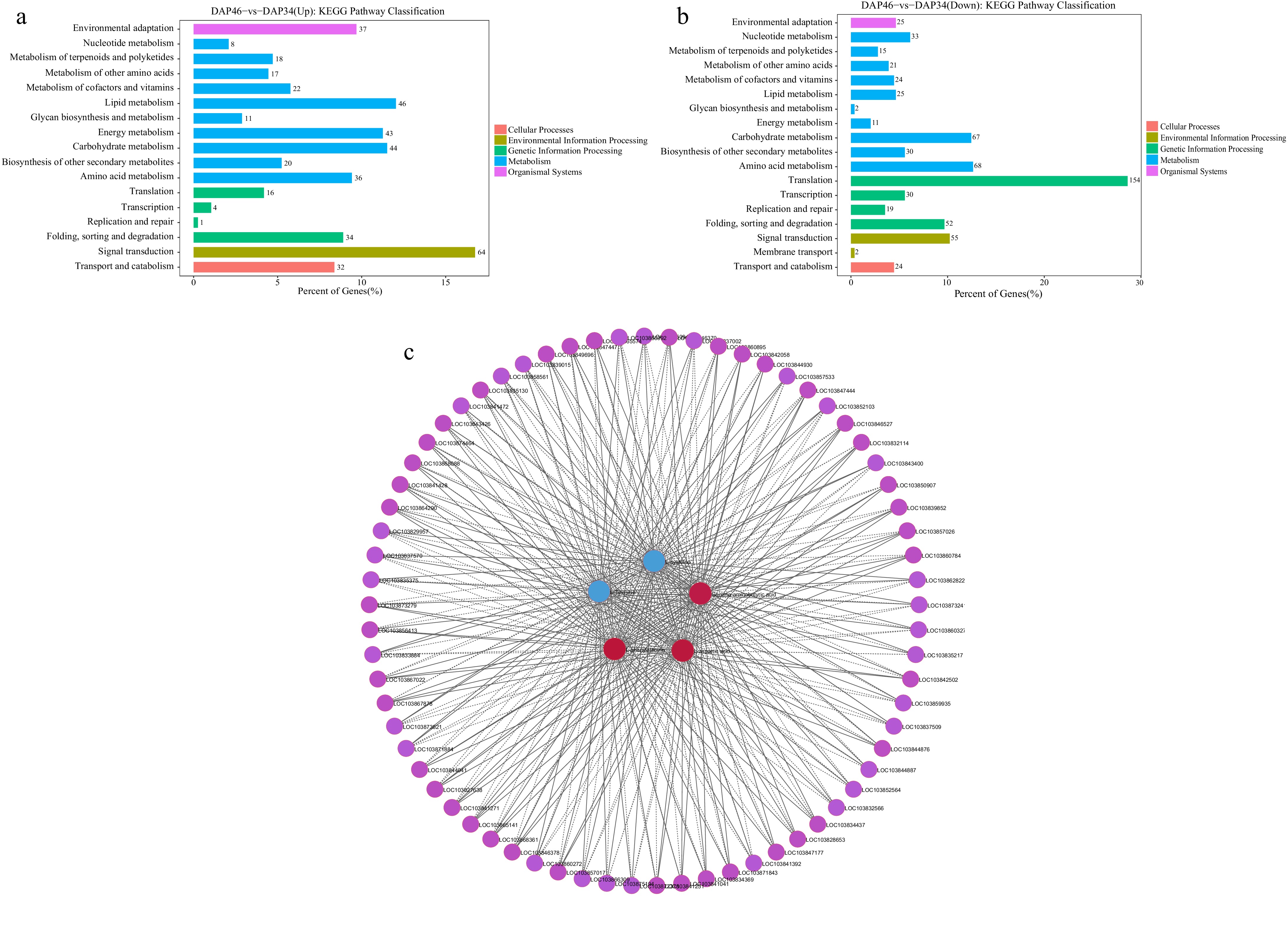
Figure 3.
Screening of genes related to key amino acids. KEGG pathway classification of (a) upregulated, and (b) downregulated genes. (c) Correlation network diagram illustrating between the relationships between five key amino acids and the top 500 genes with Fragments Per Kilobase of transcript per Million mapped reads (FPKM) value where p < 0.05.
-
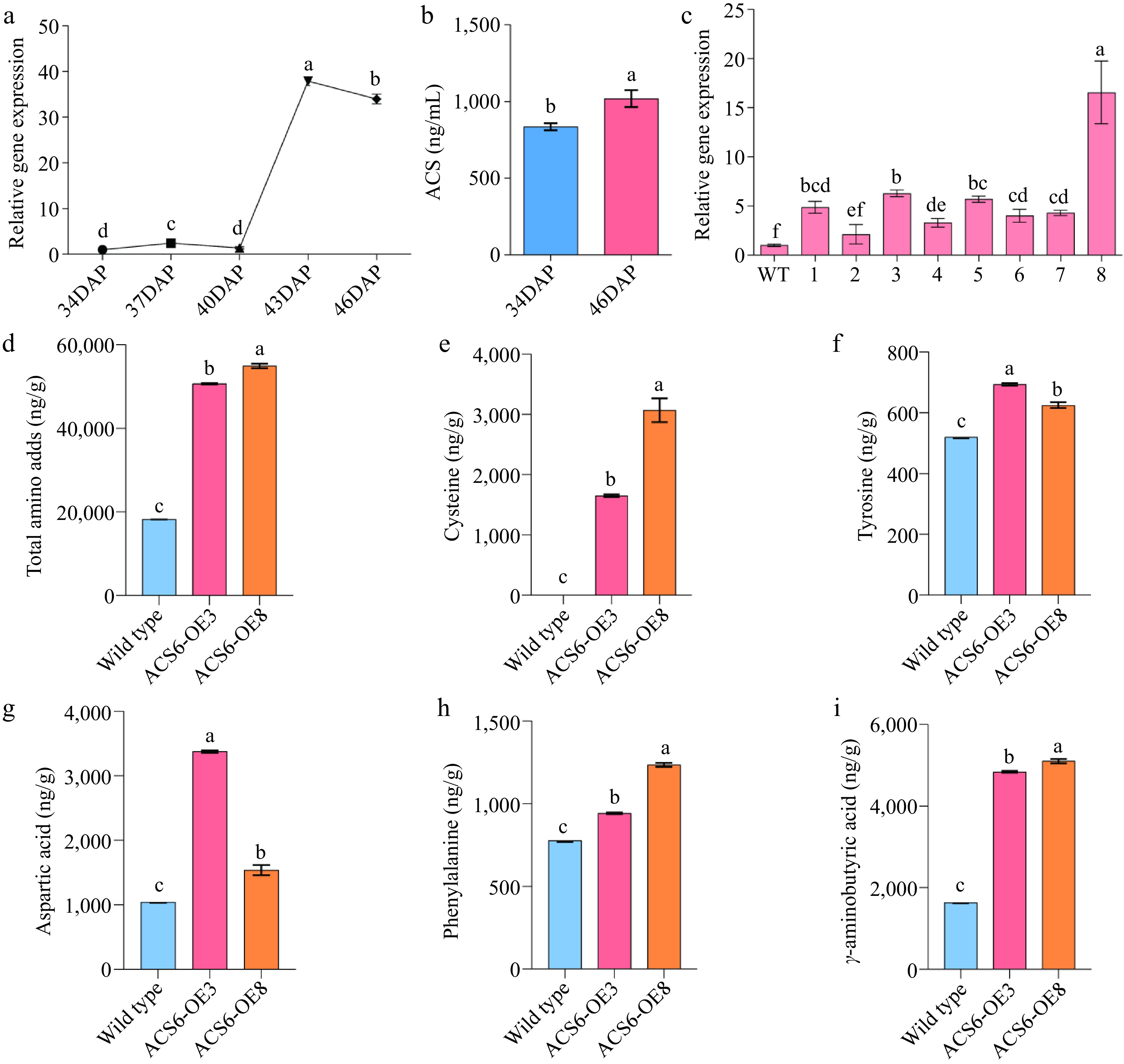
Figure 4.
Verification of BcACS6 overexpression. (a) Relative expression levels of the BcACS6 gene at different developmental stage. The relative expression level at 34 days after planting (DAP) was normalized to 1. (b) Assay results of 1-aminocyclopropane-1-carboxylic acid synthase (ACS) enzyme activity. (c) Relative expression levels of BcACS6 in T2 transgenic Arabidopsis thaliana. (d)−(i) Amino acid content analysis in wild-type and BcACS6-overexpressing Arabidopsis leaves.
-
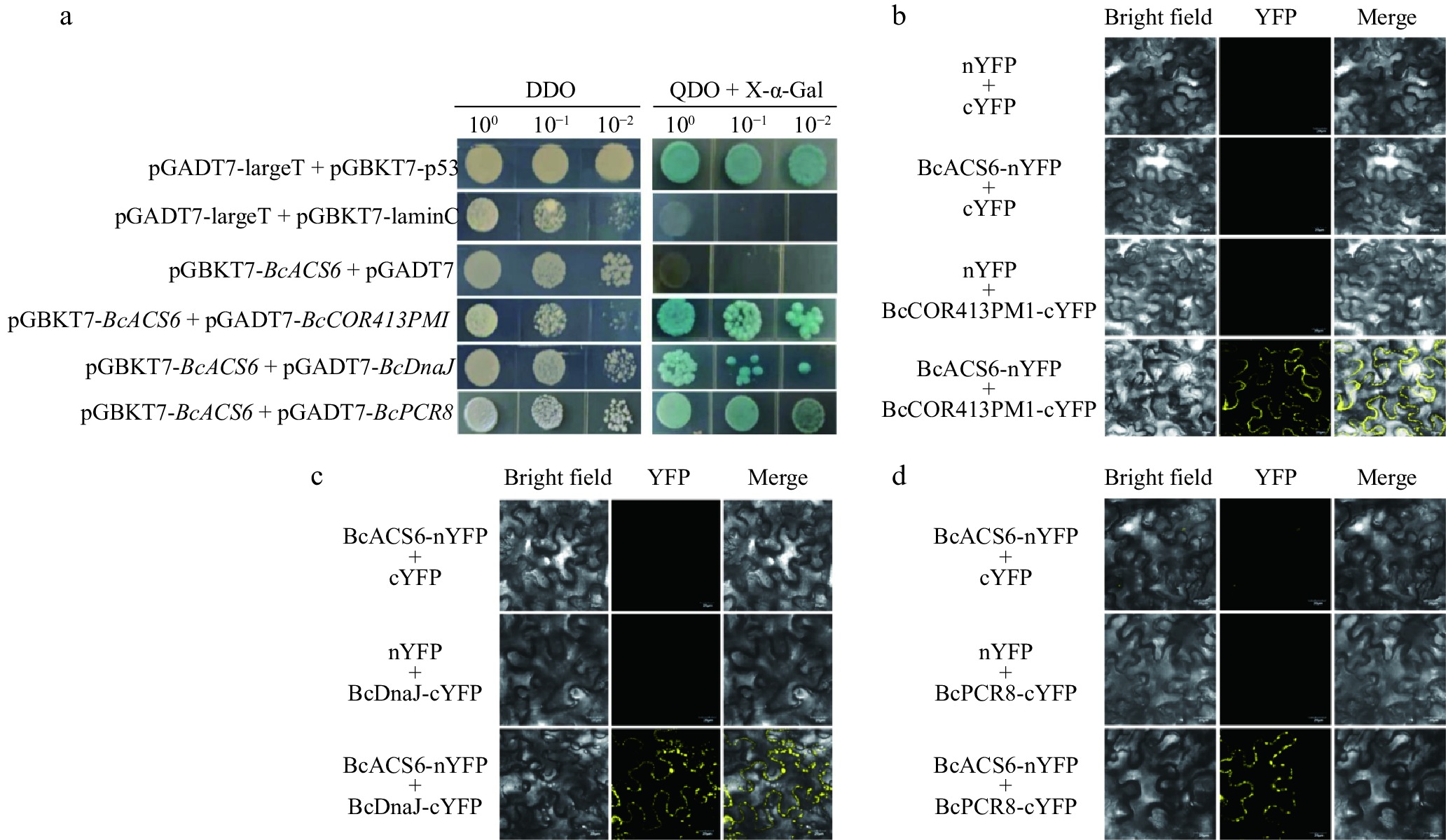
Figure 5.
Yeast two-hybrid and bimolecular fluorescence complementation (BiFC) verification. (a) Self-activation detection of the decoy vector and interaction protein verification. The positive control consisted of pGBKT7-53 + pGADT7-T, the negative control included pGADT7-Lam + pGADT7-T, and the self-activation verification group used pGBKT7-BcACS6 + pGADT7. DDO, SD medium lacking tryptophan and leucine (SD/-Trp/-Leu); QDO + X-α-gal, SD medium lacking tryptophan, leucine, histidine, and adenine (SD/-Trp/-Leu/-His/-Ade), supplemented with X-α-gal and aureobasidin A (AbA). Transformed yeast cells were spotted at 10−1 dilution on selective media. (b) BiFC assay showing interaction between BcACS6 and BcCOR413PM1 in Nicotiana benthamiana leaves. (c) BiFC assay showing interaction between BcACS6 and BcDnaJ in N. benthamiana leaves. (d) BiFC assay showing interaction between BcACS6 and BcPCR8 in N. benthamiana leaves. BcACS6, BcCOR413PM1, BcDnaJ, and BcPCR8 were fused with either the C- or N-terminus of yellow fluorescent protein (designated as cYFP or nYFP, respectively). Bright field, white light; YFP, YFP fluorescence; merge, combined signal of YFP and white light.
Figures
(5)
Tables
(0)