-
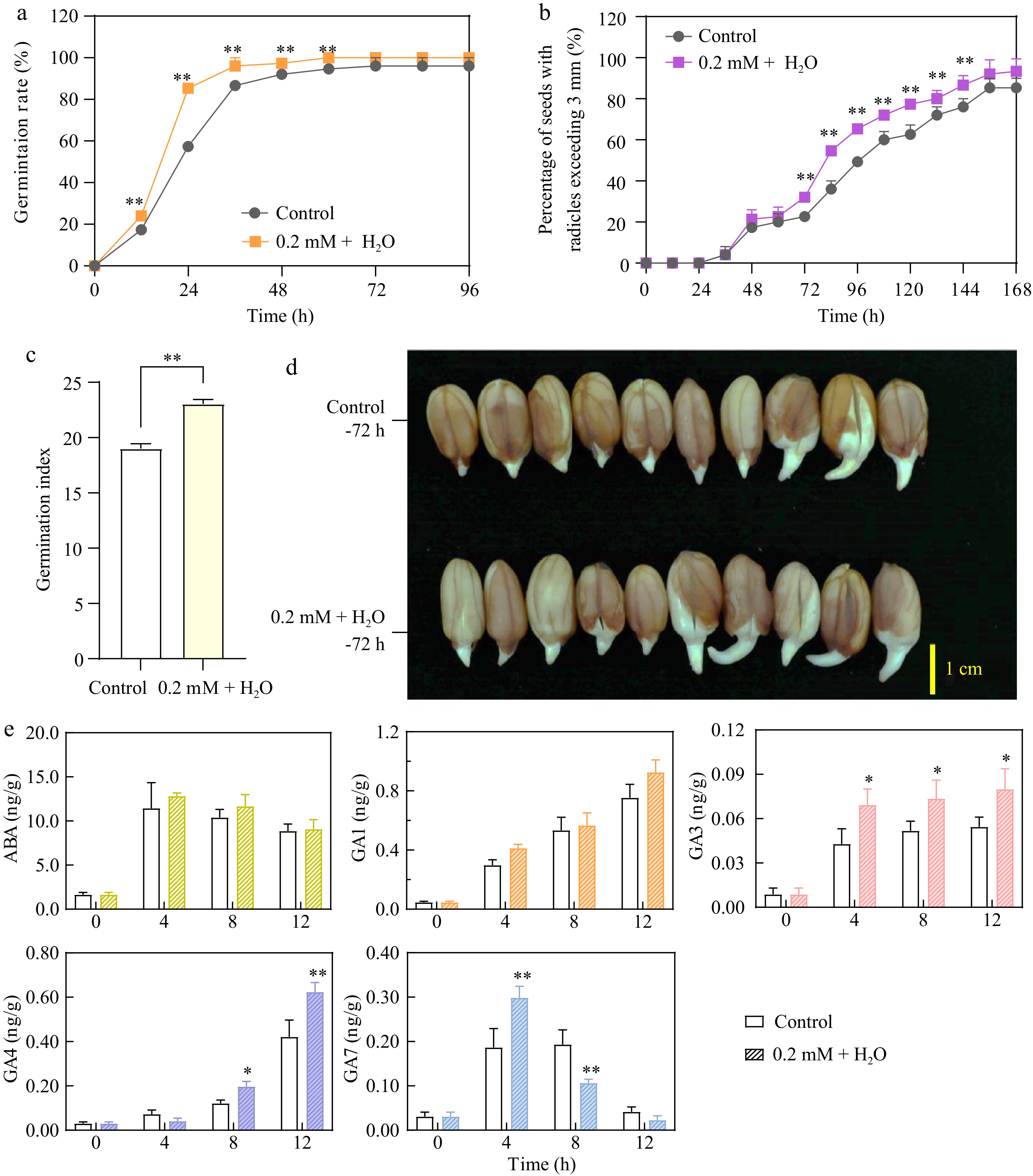
Figure 1.
0.2 mM ethylene + H2O enhances peanut seed germination, radicle growth, and gibberellin synthesis. (a) Comparison of germination rates between the control and 0.2 mM Ethylene + H2O treatment. (b) Percentage of seeds with radicles exceeding 3 mm in the control and 0.2 mM Ethylene + H2O treatment. (c) Comparison of germination indexs between the control and 0.2 mM Ethylene + H2O treatment. (d) Radicle growth at 72 h (third day) between the control and 0.2 mM Ethylene + H2O treatment. (e) Concentrations of abscisic acid and gibberellins in the control compared to the 0.2 mM Ethylene + H2O treated samples. *, p-value ≤ 0.05, **, p-value ≤ 0.01.
-
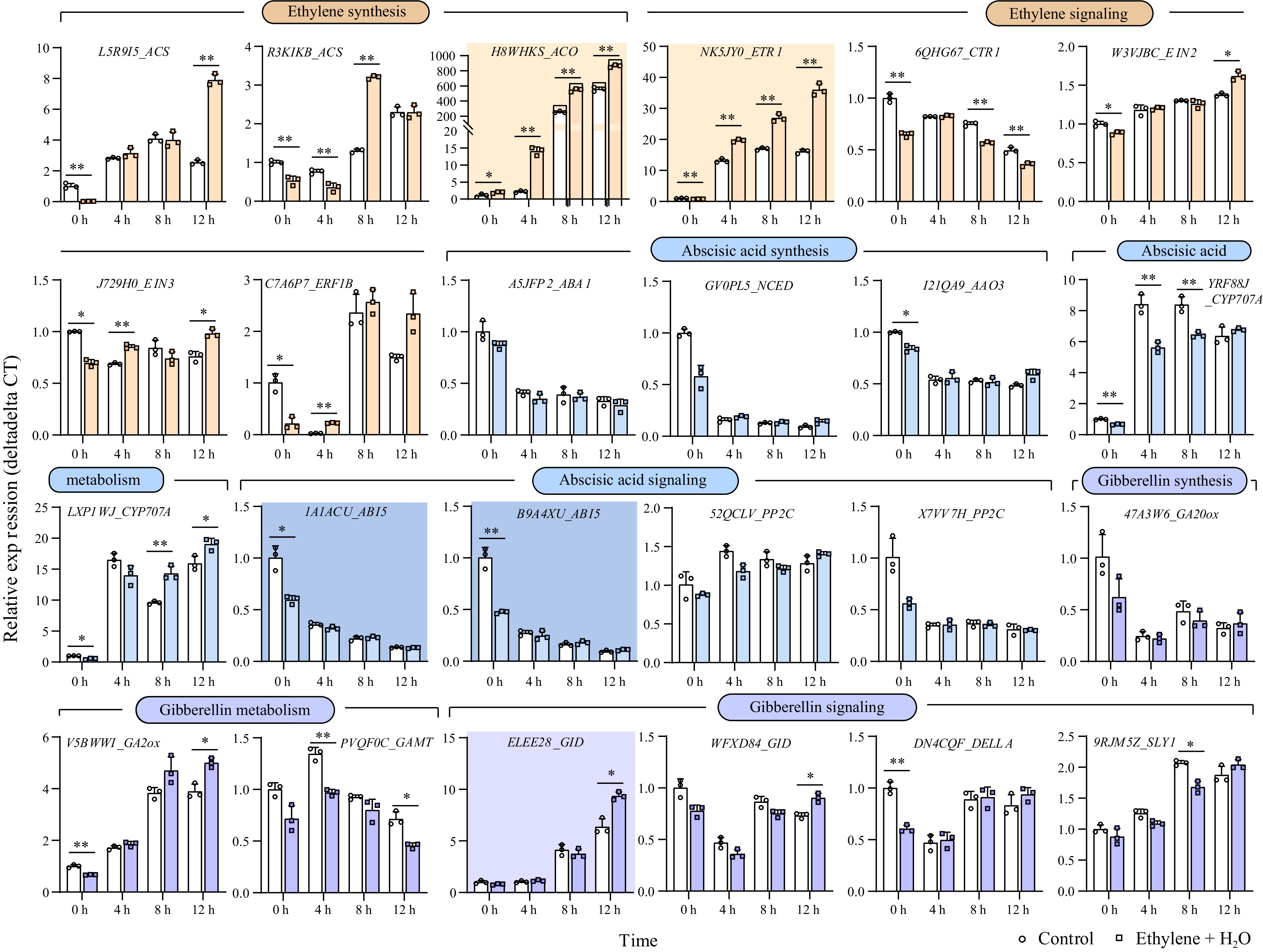
Figure 2.
Gene expression changes of key genes involved in the synthesis, degradation, and signaling of ethylene, ABA, and GA. During the first 12 h of germination, genes exhibiting more than a tenfold change in expression are highlighted with a darker background. *, p-value ≤ 0.05, **, p-value ≤ 0.01. ACS: 1-Aminocyclopropane-1-Carboxylate Synthase; ACO:1-Aminocyclopropane-1-Carboxylate Oxidase; ETR1: Ethylene Response Sensor 1; CTR1: Constitutive Triple Response 1; EIN2: Ethylene Insensitive 2; EIN3: Ethylene Insensitive 3; ERF1B: Ethylene Response Factor 1B; ABA1: Zeaxanthin Epoxidase; NCED: 9-cis-epoxycarotenoid dioxygenase; AAO3: Abscisic Aldehyde Oxidase 3; CYP707A: Cytochrome P450, Family 707, Subfamily A; ABI5: Abscisic Acid Insensitive 5; PP2C: Protein Phosphatase 2C; GA20ox: Gibberellin 20 Oxidase; GA2ox: Gibberellin 2 Oxidase; GAMT: Gibberellin Methyltransferase; GID: Gibberellin Insensitive Dwarf; DELLA: Aspartic acid (D), Glutamic acid (E), Leucine (L), Leucine (L), and Alanine (A)—in a conserved region of the protein; SLY1: SLEEPY1.
-
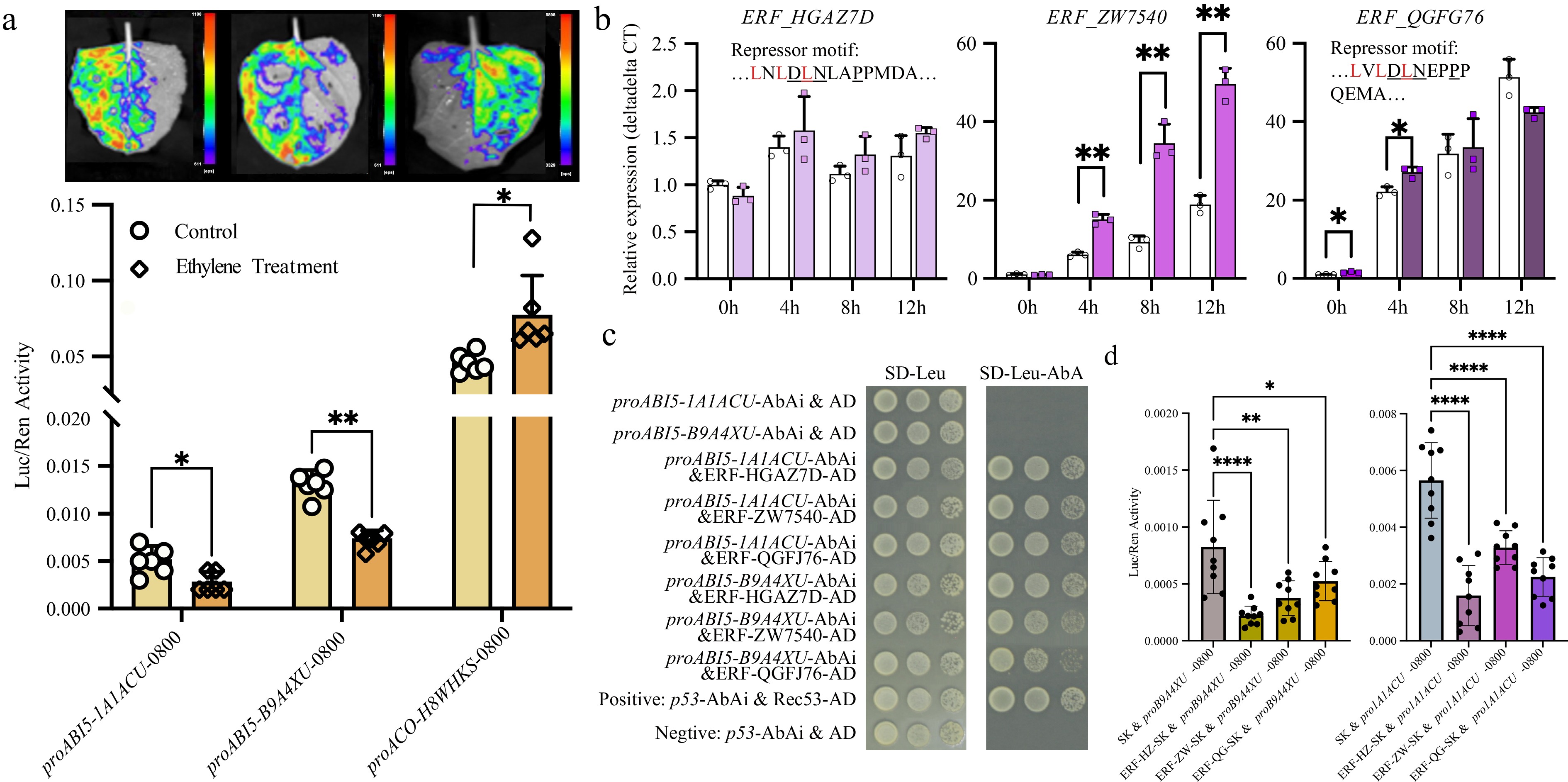
Figure 3.
Ethylene inhibits ABI5 expression through ERFs. (a) Dual-luciferase assays confirmed that ethylene activated ACO expression while suppressing ABI5 expression. (b) Gene expression changes of three ERF genes. Two of them (arahy.HGAZ7D and arahy.QGFJ76) contained repressor motifs. (c) Yeast one-hybrid assays confirmed that all three ERFs bound to the promoter of ABI5. (d) Dual-luciferase assays confirmed that all three ERFs inhibited ABI5 expression. *, p-value ≤ 0.05; **, p-value ≤ 0.01.
-
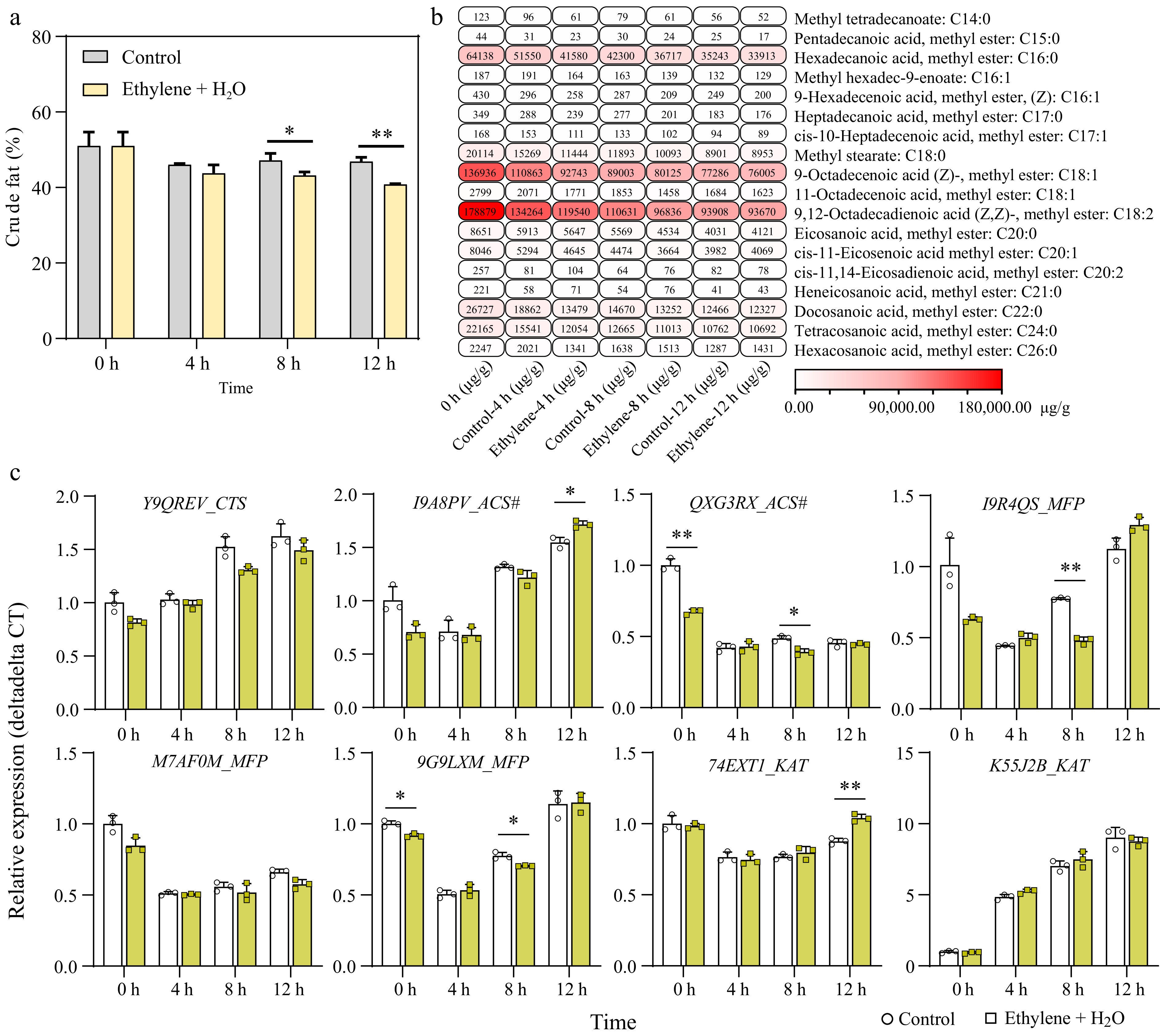
Figure 4.
Ethylene enhances fatty acid degradation during seed germination. (a) Change in crude fat concentration between the control and 0.2 mM Ethylene + H2O treatment during germination. (b) GC-MS (Gas Chromatography-Mass Spectrometry) analysis of fatty acid composition changes in samples between control and ethylene treatment, the detailed data can be found in Supplementary Table S3 and Supplementary Fig. S3. (c) Expression changes of key genes in the beta-oxidation pathway during seed germination, comparing control and ethylene treatment. *, p-value ≤ 0.05; **, p-value ≤ 0.01. CTS: Comatose/Peroxisomal ABC Transporter; ACS#: Acyl-CoA Synthetase. #, not to be confused with the ethylene-related ACS; MFP: Multifunctional Protein–Contains 2E-enoyl-CoA hydratase and 3S-hydroxyacyl-CoA dehydrogenase activities; KAT: 3-Ketothiolase.
-
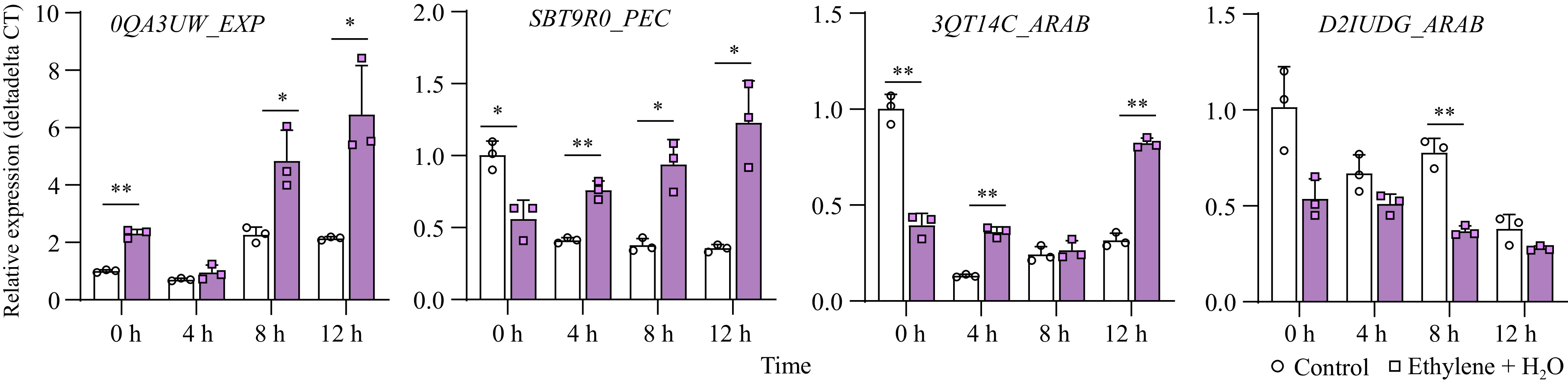
Figure 5.
Expression changes of cell wall-loosening related genes between control and ethylene treatment. *, p-value ≤ 0.05; **, p-value ≤ 0.01. EXP: Expansin; PEC: Pectinesterase; ARAB: arabinogalactan protein.
-
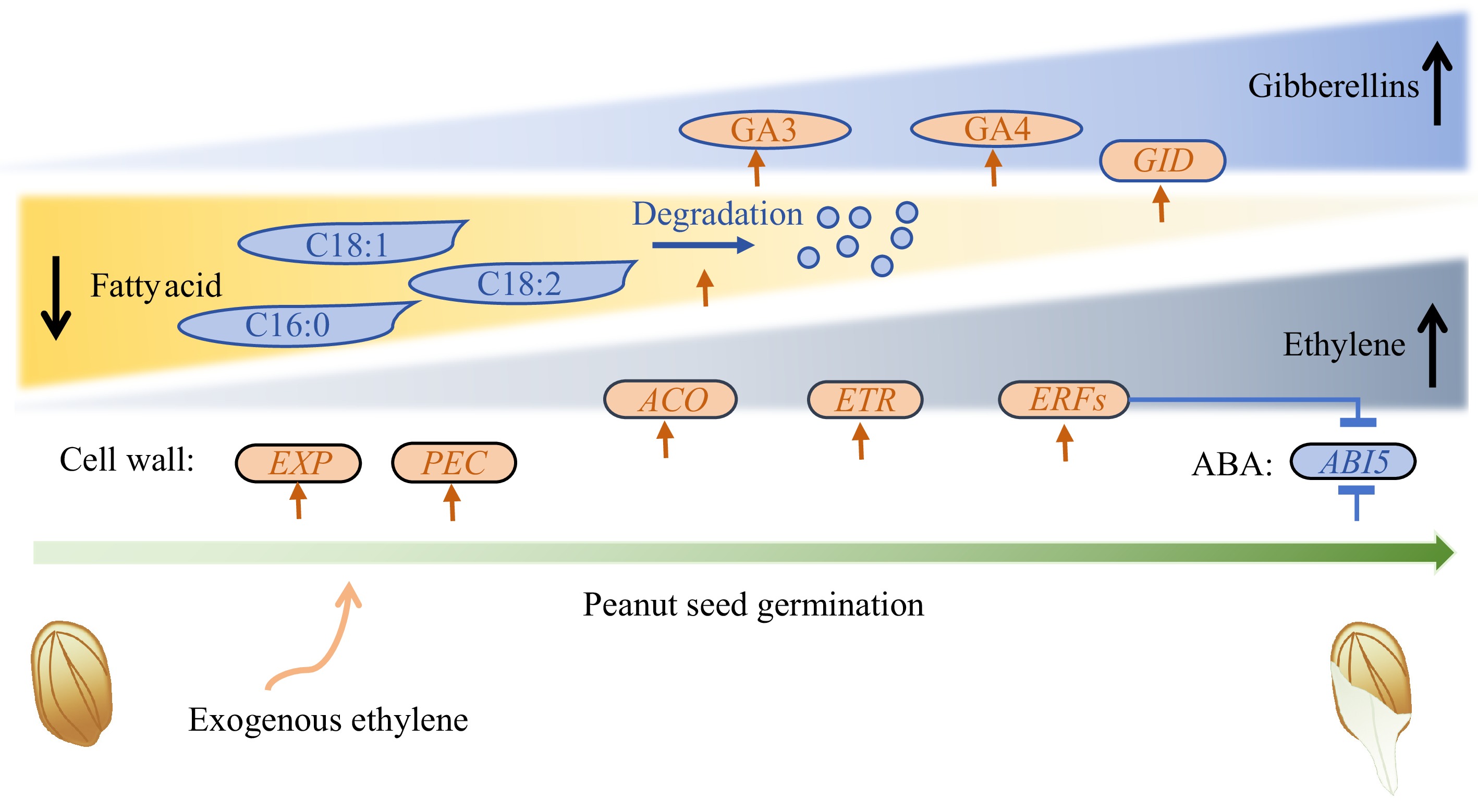
Figure 6.
Schematic depiction of ethylene's effect on peanut seed germination. Round rectangles represent genes, ovals represent plant hormones, and shapes with sharp corners represent fatty acid. EXP: Expansin; PEC: Pectinesterase; ABI5: Abscisic Acid Insensitive 5; GID: Gibberellin Insensitive Dwarf; ACO: 1-Aminocyclopropane-1-Carboxylate Oxidase; ETR: Ethylene Response Sensor; ERF: Ethylene Response factor.
Figures
(6)
Tables
(0)