-
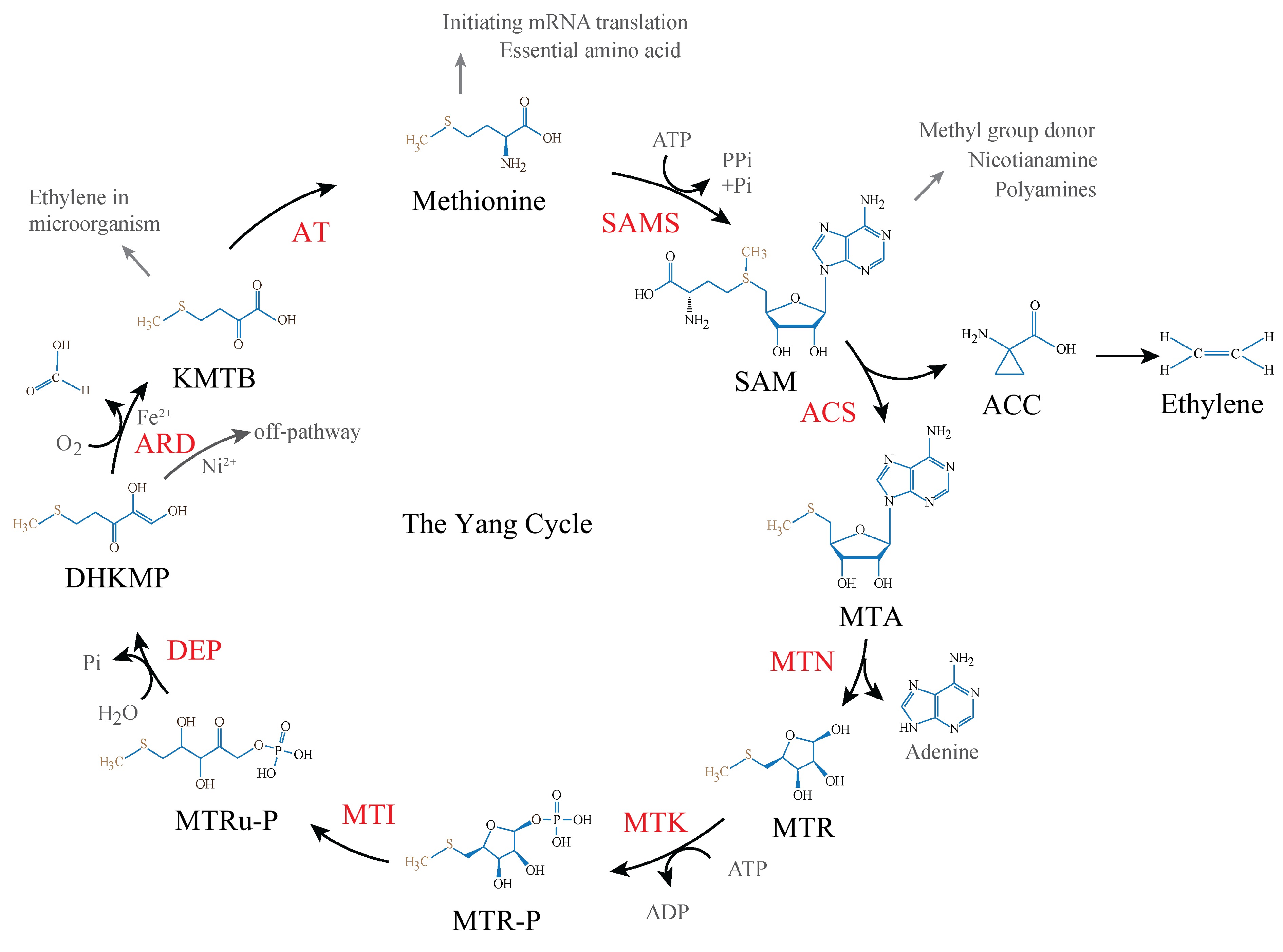
Figure 1.
The Yang cycle in plants. S-adenosyl-methionine (SAM) is first derived from methionine by SAM synthetase (SAMS), SAM then is converted to 5'-S-methyl-5'-thioadenosine (MTA) and 1-aminocyclopropane-1-carboxylic acid (ACC) by ACC synthase (ACS). MTA is then depurinated to 5-methylthioribose (MTR) by MTA nucleosidase (MTN). MTR is subsequently phosphorylated to 5-methylthioribose-1-phosphate (MTR-P), catalyzed by MTR kinase (MTK) in the presence of adenosine triphosphate (ATP). MTR-P gets isomerization to yield 5-methylthioribulose-1-phosphate (MTRu-P) by MTR-P isomerase (MTI). MTRu-P then is metabolized to 1,2-dihidroxy-3-keto-5-methylthiopentene (DHKMP) by dehydratase-enolase-phosphatase (DEP). DHKMP is converted to 2-keto-4-methylthiobutyrate (KMTB) by acireductone dioxygenase (ARD) in the penultimate step. At last, KMTB turns to methionine by an unknown aminotransferase (AT).
-
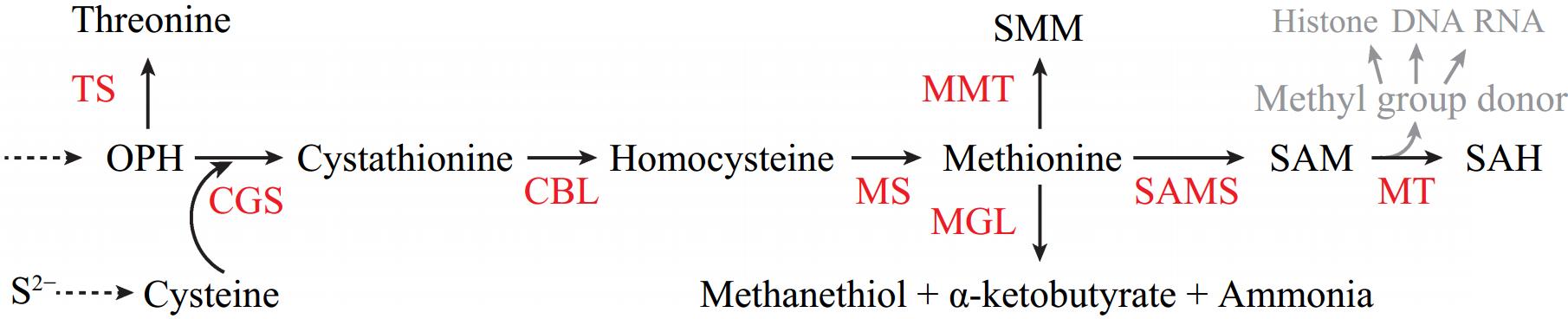
Figure 2.
The metabolism and homeostasis of methionine and S-adenosyl-methionine (SAM) in plants. Methionine can be de novo synthesized from O-phosphohomoserine (OPH), in which cystathionine γ-synthase (CGS) competes with threonine synthase (TS) for OPH. CGS metabolizes OPH and cysteine to cystathionine which is then converted to homocysteine by cystathionine β-lyase (CBL). Methionine is produced from homocysteine by methionine synthase (MS). Methionine can be metabolized to methanethiol, α-ketobutyrate and ammonia by methionine γ-lyase (MGL). It also can be converted to S-methylmethionine (SMM) by methionine S-methyltransferase (MMT). The conversion of SAM to S-adenosylhomocysteine (SAH) by diverse methyltransferases (MTs) provides methyl moiety for acceptors including histone, DNA, and RNA.
-
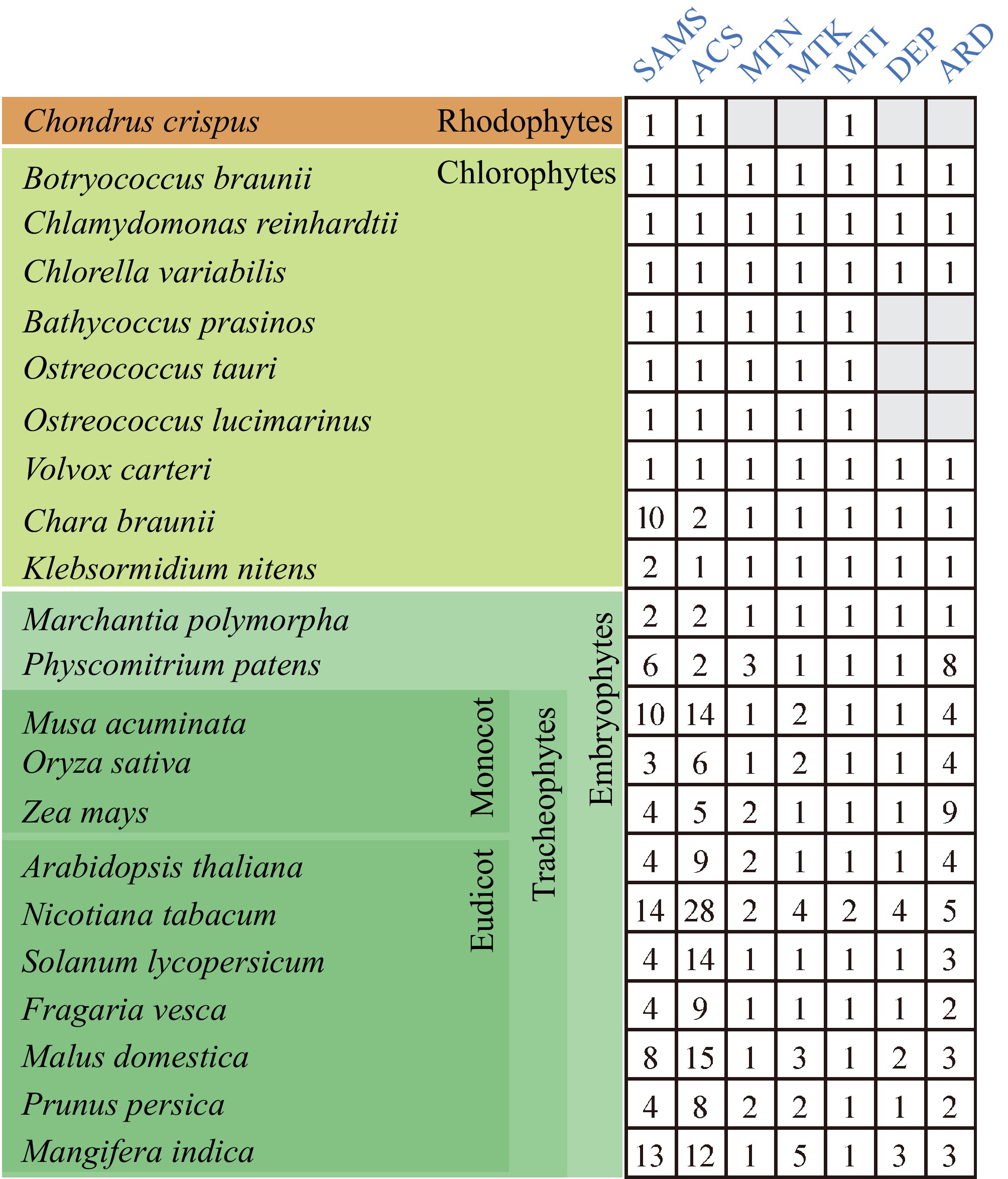
Figure 3.
The enzyme proteins of the Yang cycle in several major plant lineages. The Yang cycle proteins in each species were identified using BLAST with an E-value threshold of 1e−5, employing the default settings of TBtools-II. Searches were conducted with the amino acid sequences of each enzyme from Arabidopsis thaliana as references. The numbers indicate enzyme paralogs. Gray boxes indicate an absence of enzyme paralog.
-
Enzyme Plant species Main content Ref. SAMS Arabidopsis thaliana Excessive methionine accumulates in the mto3-1 and mto3-2 mutants [61,62] Overexpressing SAMS is morphologically indistinguishable from wild-type plants, or leads to abnormal floral organ development [62,65] SAMS is phosphorylated by CDPK28 [74] Nicotiana tabacum Suppressing SAMS renders accumulation of methanethiol and methionine [63] Rice Suppressing OsSAMS1, 2 and 3 causes pleiotropic phenotypes [64] OsSAMS1 functions as a regulator for grain size and yield [67] OsWAK112 interacts with OsSAMS1, 2 and 3 [72] OsSAMS1 interacts with OsLCD3 [76] OsSAMS1 is targeted by F-box protein OsFBK12 [77] Pumpkin SAMS interact with a long non-coding RNA with promoted stability [76] Tomato SlSAMS1 influences fruit ripening and it interacts with FERONIA-like [79] ACS Arabidopsis thaliana The expression of ACS genes are not vascular-specific [6,83] ACS has an additional Cβ-S lyase activity [84] MTN Lupinus luteus Enzyme is purified from seed [86] Arabidopsis thaliana MTN1 and MTN2 exhibit diverse expression patterns, preferentially in vascular tissues [6] The cystal structures of MTN1 and MTN2 are determined [88,89] mtn1 and mtn2 single and double mutants are characterized [42,43] Rice OsMTN recombinant enzyme is characterized [90] Maize ZmMTN1 mutant is linked with Fe and NA hemeostasis [91] Apple MTN activity is first detected in fruit extract [85] Tomato SlMTN expression and activity are characterized in fruit [28,48] MTK Lupinus luteus MTK activity is partially purified from seed [93] Rice OsMTK1 and OsMTK2 are cloned and evaluated under sulfur deficiency [94] Arabidopsis thaliana MTK is preferentially expressed in phloem [6] MTK is cloned and T-DNA insertional mutants are characterized [94] An eto3mtk double mutant is evaluated [95] Tomato SlMTK expression and activity are characterized in fruit [28,48,96] MTI Arabidopsis thaliana MTI is cloned and shows phloem-specific in expression [6] The functions of MTI in sulfur metabolism during flowering and seed development is evaluated [99] DEP Arabidopsis thaliana DEP is cloned and its expression also shows phloem-specific [6] The functions of DEP in sulfur metabolism during flowering and seed development is evaluated [99] Apple MdDEP1 is ectopically expressed in Arabidopsis, enhancing stress tolerance and flowering [100] The expression of MdDEP1 is regulated by MdBHLH3, and the activity is affected by MdY3IP1 [101,102] ARD Rice ARD has dual enzymatic activity with different binding metals, OsARD1 is induced by submergence and ethylene [107] OsARD1 promotes stress tolerance [110] Potato StARD1 is wounding responsive [109] Arabidopsis thaliana ARD1 function in hypocotyl growth is evaluated, ARD1 is an effector for AGB1 [111] Tomato The expression of SlARD1 and SlARD2 is characterized in fruit [28] Apple The physiological roles of an apple ARD gene are investigated by ectopically expressing in tomato plant [113] AT Tomato and maize GTK is proposed to convert KMTB to methionine [115] Table 1.
Studies on the enzymes involved in the Yang cycle in plants.
Figures
(3)
Tables
(1)