-
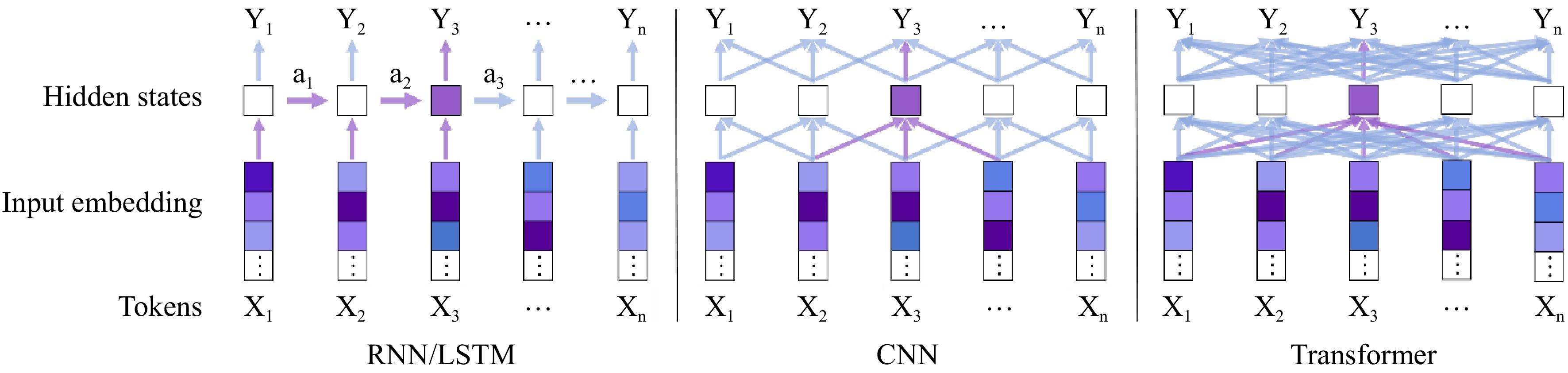
Figure 1.
Comparison of deep learning sequence models[49]: RNN / LSTM: iteratively transmitting sequence information based on hidden states. CNN: Converging the data of adjacent areas through the local perceptual field of view. Transformer: The self-attention mechanism is used to fully capture the pattern information of any span of the input sequence.
-
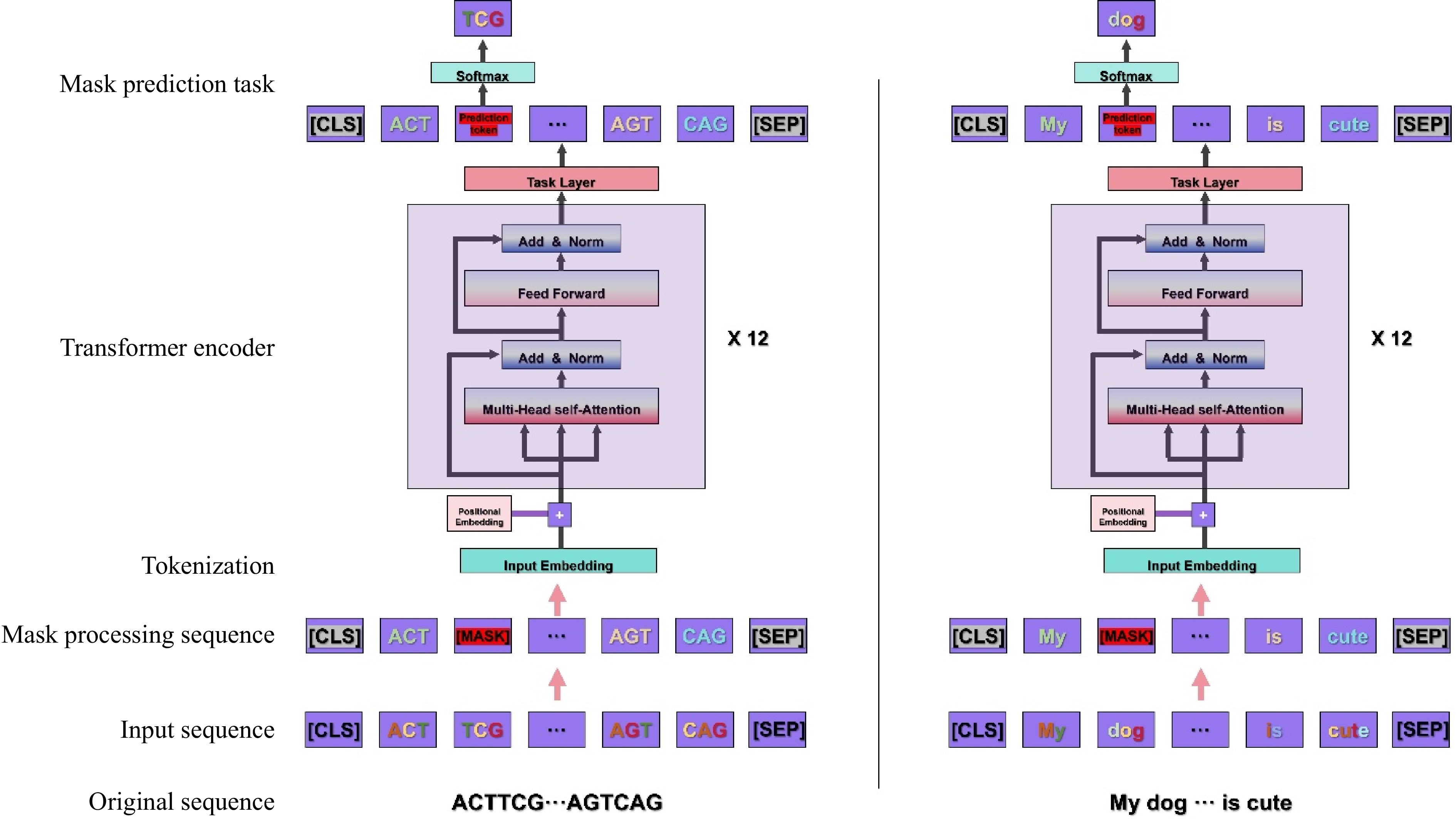
Figure 2.
Illustration of the masked pre-training task process for BERT models based on DNA and text sequences[49,51]. The process is as follows: 1) Input sequence preparation: One-dimensional character sequences serve as the initial input data. 2) Data preprocessing: The original character sequences undergo segmentation, trimming, and padding to standardize the sequence length, ensuring a consistent data format for subsequent processing. 3) Tokenization, masking, and special token insertion: The preprocessed sequences are tokenized, transforming character sequences into token sequences. Tokens within the sequence are randomly selected for masking, which involves replacing them with a special [MASK] token, randomly substituting them with other tokens, or leaving them unchanged. Special tokens [CLS] and [SEP] are inserted at the beginning and end of the sequence, respectively, to denote the start and end of the sentence, aiding the model in comprehending the overall structure of the sequence. Tokens, as the fundamental units for model learning, encode the semantic information of the sequence. 4) Embedding layer processing: The tokenized sequences are fed into the embedding layer, where each token is converted into a vector representation, thereby extracting and encoding the feature information of the data. 5) Transformer module training: The embedded vector sequences are input into the Transformer module for deep training. The Transformer module comprises multiple stacked encoder layers, each containing components such as self-attention mechanism, feedforward neural network, and normalization layer. These components collectively process the sequence to capture linguistic features and contextual information. 6) Model output and token prediction: The model outputs the vector representation of the masked tokens and attempts to predict the original tokens at the masked positions. 7) Similarity computation and parameter updating: The similarity between the predicted tokens and the original (pre-masked) tokens is computed (typically using the cross-entropy loss function). Through the backpropagation algorithm, the loss information is propagated back to the model, thereby updating the model parameters and gradually enhancing the model's ability to predict masked tokens. 8) Training iteration and optimization: The process is repeated over multiple training epochs to continuously optimize the model parameters, thus continuously improving the model's understanding and generation capabilities for the sequences.
-
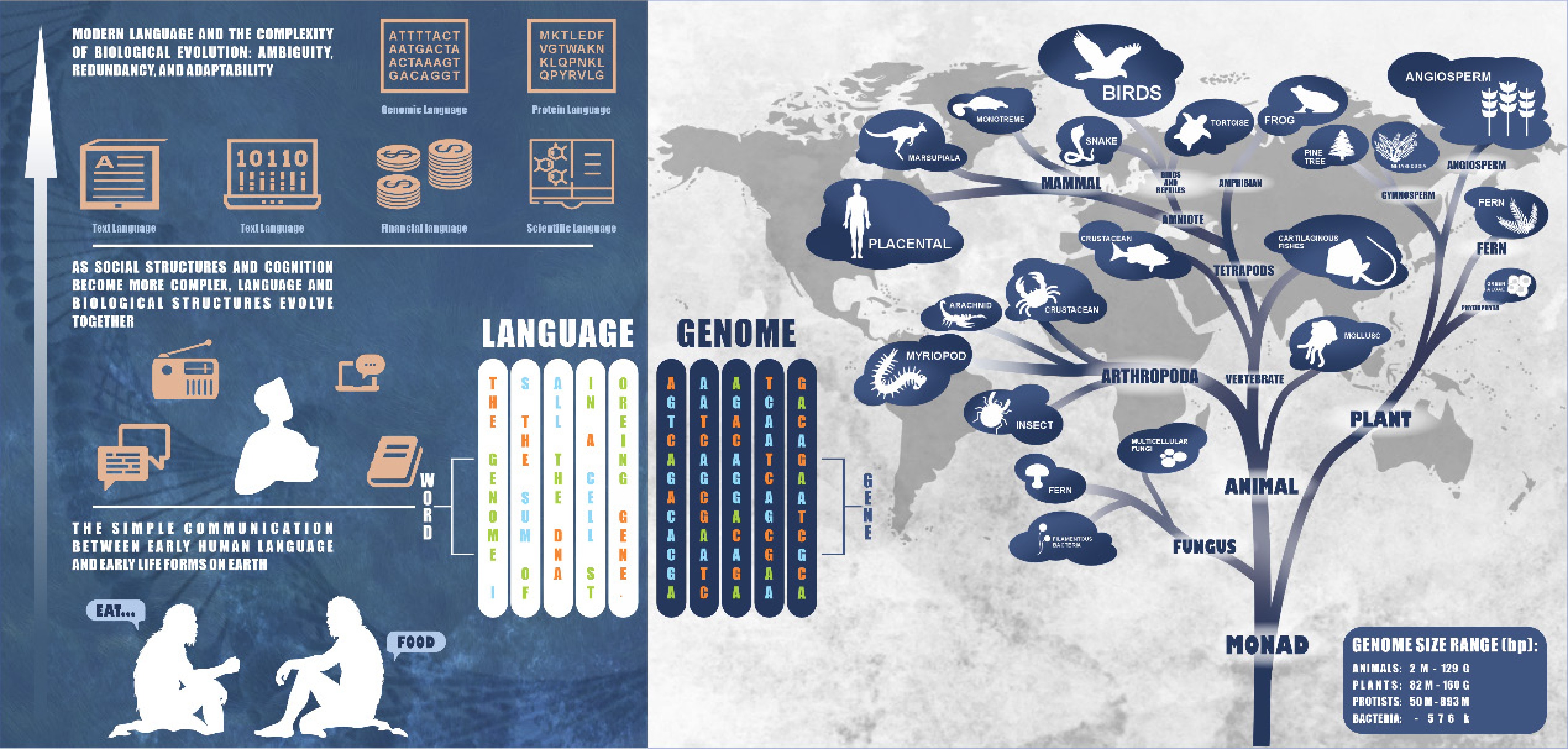
Figure 3.
Similarity between genome sequence and language sequence. The genome size data is derived from the NCBI genome database[82].
-
Model Time Base model Model architecture Parameters Pre-trained data species Capability Tokenizer type Tokens length (bp) Ref. DNABERT 2021.02 BERT Transformer Encoder 110 M Animal Function Prediction K-mer (3−6) 512 [49] Enformer 2021.10 Transformer + CNN Transformer Encoder 240 M Animal Function prediction Single-base 196,608 [81] MoDNA 2022.08 BERT Transformer encoder - Animal Function prediction K-mer (6) 512 [84] GPN 2022.08 BERT + CNN Transformer encoder − Plant Variant prediction Single-base 512 [33] iEnhance-BERT 2022.08 BERT(DNABERT) +
CNNTransformer encoder − − Enhancer prediction K-mer (3−6) 512 [85] FloraBERT 2022.08 BERT Transformer encoder − Plant Function prediction BPE 256 [36] TFBert 2022.09 BERT(DNABERT) Transformer Encoder 110 M Animal Epigenomics prediction K-mer (3−6) 512 [86] iDNA-ABF 2022.10 BERT(DNABERT) Transformer encoder 110 M Animal Function prediction K-mer (3−6) 512 [24] iEnhancer-ELM 2022.12 BERT(DNABERT) Transformer encoder 110 M Animal Enhancer prediction K-mer (3−6) 512 [87] Species-aware DNALM 2023.01 BERT(DNABERT) Transformer encoder 110 M Fungus Function prediction K-mer (6) 512 [25] Nucleotide Transformer 2023.01 BERT Transformer encoder 50 M−2.5 B Animal Function Prediction K-mer (6) 1,000−2,048 [88] GPN-MSA 2023.01 BERT Transformer encoder 86 M Animal Variant prediction Single-base 128 [89] SpliceBERT 2023.02 BERT Transformer encoder 19.4 M Animal Splicing prediction Single-base 1,024 [28] miProBERT 2023.03 BERT(DNABERT) Transformer encoder 110 M − Promoter prediction K-mer (6) 512 [90] RNA-MSM 2023.03 BERT+CNN Transformer encoder − − Structure prediction Single-base 512 [31] MRM-BERT 2023.06 BERT(DNABERT) Transformer encoder 110M Animal + Microorganism RNA modifications prediction K-mer (6) 512 [26] GENA-LM 2023.06 BERT/BigBird Transformer encoder 110−360 M Animal Function prediction BPE 512−4,096 [91] DNABERT-2 2023.06 BERT Transformer encoder 117 M Animal Function prediction BPE 128 [92] Geneformer 2023.06 BERT Transformer encoder − Animal Function prediction Custom tokenizer 2,048 [93] PLPMpro 2023.07 BERT(DNABERT) Transformer encoder − Eucaryon Promoter prediction K-mer (6) 512 [94] EpiGePT 2023.07 BERT + CNN Transformer encoder − Animal Epigenomics prediction Single-base 128 [29] Uni-RNA 2023.07 BERT Transformer encoder 25−400 M − Function prediction Single-base 512−1,280 [32] AgroNT 2023.10 BERT Transformer encoder 1B Plant Function prediction K-mer (6) 1,024 [35] FGBERT 2024.02 BERT Transformer encoder 954.73 M Metagenomics Functional prediction − − [27] RiNALMo 2024.02 BERT Transformer encoder 135−650 M − Functional prediction Single-base 1,024 [95] gLM 2024.04 BERT Transformer encoder 1B Metagenomics Functional prediction − − [96] RNAErnie 2024.05 BERT Transformer encoder 105 M − Functional prediction Single-base 512 [30] GenSLMs 2022.10 Diffusion + GPT Transformer decoder 25−250 M Virus Variant prediction K-mer (3) 2,048 [97] DNAGPT 2023.08 GPT Transformer decoder 100 M−3 B Animal Function prediction + Sequence generation K-mer (6) 512−4,096 [98] ENBED 2023.11 Transformer Transformer encoder−decoder 1.2 B Animal + Plant + Insect + Bacteria Function prediction + Sequence generation Single-base 16,384 [99] HyenaDNA 2023.06 Hyena Hyena 0.44−6.6 M Animal Function prediction + Species classfication Single-base 64 K−1 M [100] Evo 2024.02 StripedHyena StripedHyena 7B Bacteria + archaea Function prediction + Sequence generation Single-base 131 K [101] PDLLM 2024.12 Mamba Mamba 130 M Plant Function prediction Single-base/
K-mer (2−6)/BPE512 [34] Table 1.
Summary table of genomic LLMs.
Figures
(3)
Tables
(1)