-
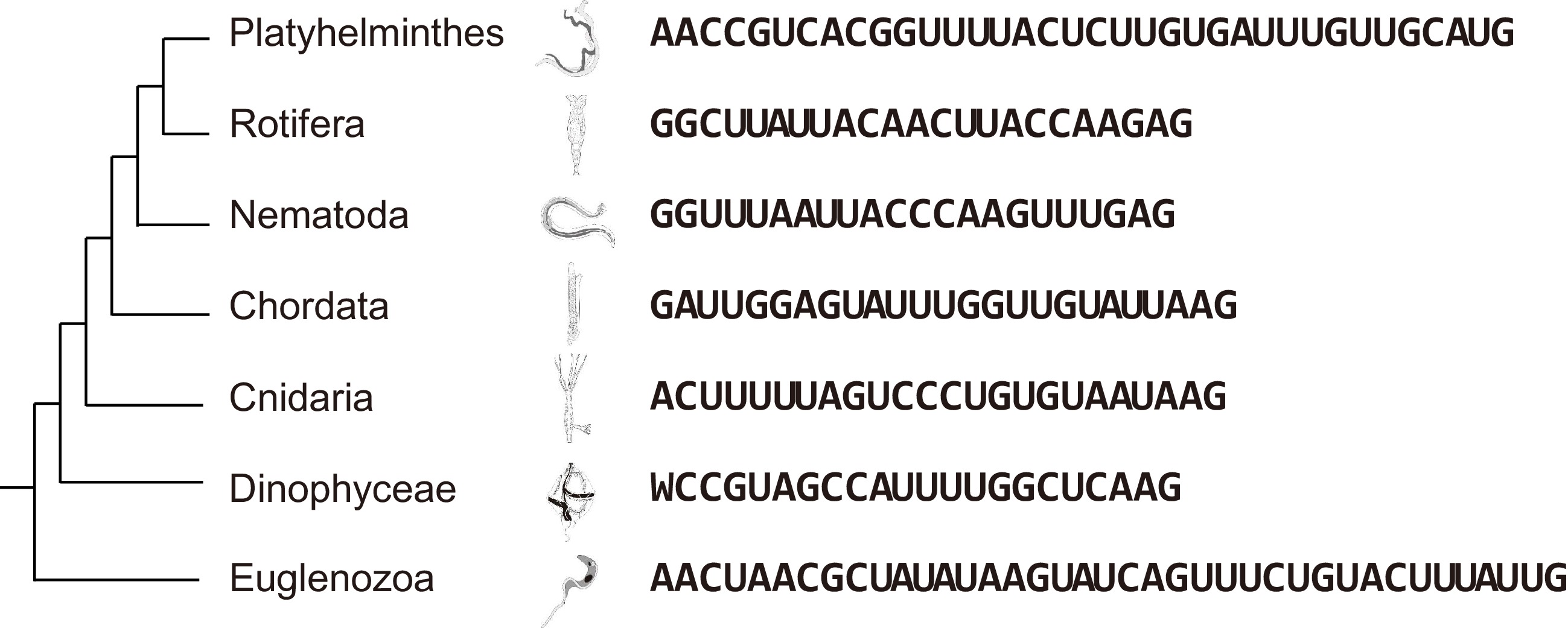
Figure 1.
SL sequences vary among different phyla but are conserved at their 3' ends.
-
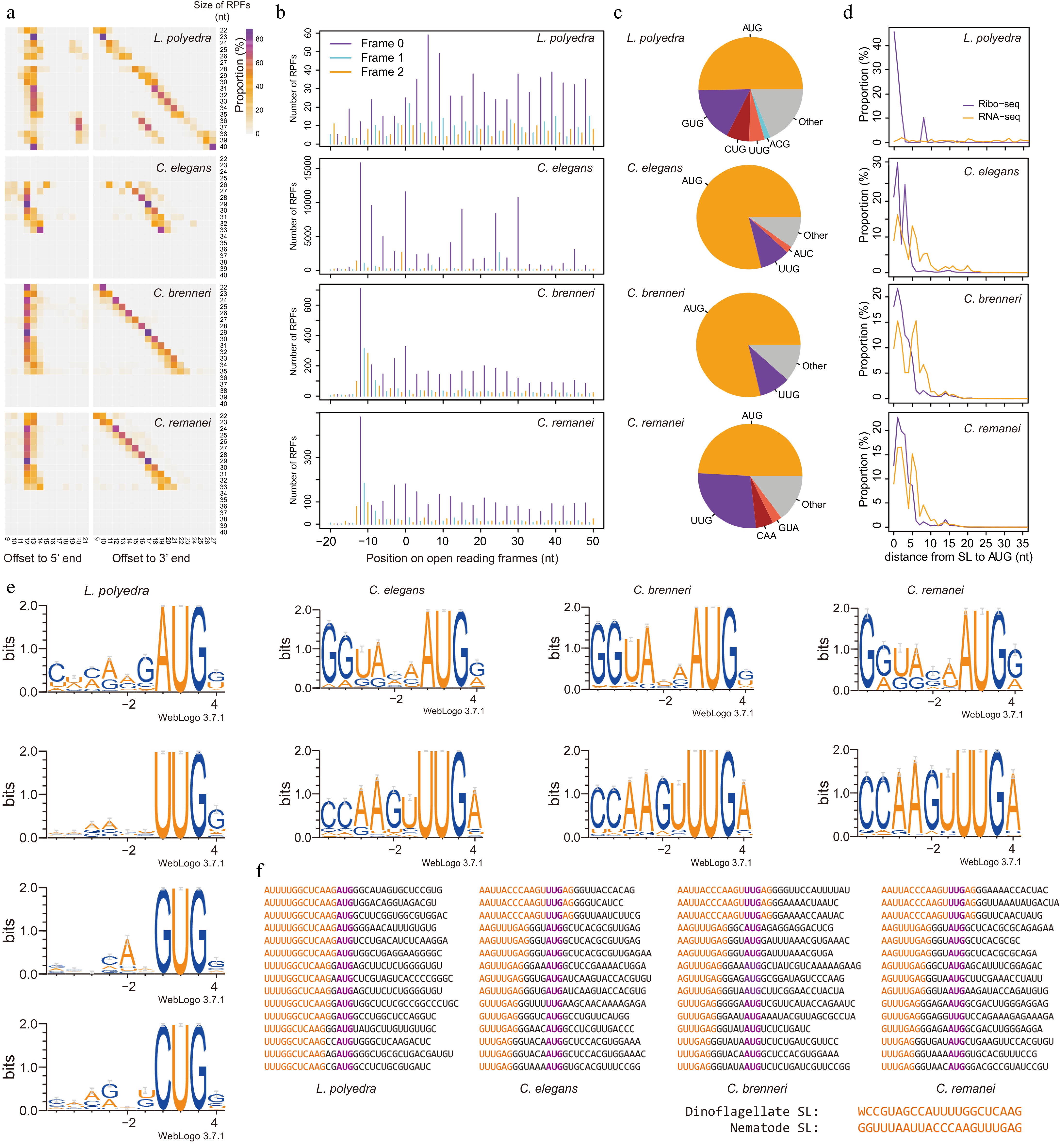
Figure 2.
SL provides Kozak consensus for the selection of start codons in dinoflagellates and nematodes. (a) Offsets from P-site to the 5' and 3' end of RPFs. All the RPFs were mapped to the genomes or transcriptome (L. polyedra) and the RPFs overlapped with the annotated start codons were used to count the offsets from their 5' and 3' ends to the annotated start codons (P-sites). (b) The mapping positions of RPFs on open reading frames display strong 3-nt periodicity, suggesting a high quality of these datasets. Frame 0 represents translating frames of genes and frame 1 and 2 represent the frames shifted 1 or 2 nt. The X-axis show the position of RPFs on the open reading frames, where position 0 represents the position of start codons. (c) The usage of start codons in different species. (d) Distance from SL to start codon in RPFs and RNA-seq reads. (e) Kozak consensus of different start codons identified from SL-RPFs. (f) Examples of SL-RPFs in dinoflagellates and nematodes. The start codon is colored in purple and the SL sequence is colored in orange.
Figures
(2)
Tables
(0)