-

Figure 1.
Heterogeneity of tea leaf metabolites in five tea cultivars. (a) Leaf amino acids. (b) Leaf catechins. (c) Other leaf secondary metabolites. A total of 19 free amino acids were determined, including theanine (THEA), alanine (ALA), arginine (ARG), asparagine (ASN), aspartic acid (ASP), γ-aminobutyric acid (GABA), glutamine (GLN), glutamic acid (GLU), histidine (HIS), Isoleucine (ILE), leucine (LEU), lysine (LYS), phenylalanine (PHE), proline (PRO), serine (SER), threonine (THR), tryptophan (TYP), tyrosine (TYR), valine (VAL), and glycine (GLY). The determined catechins included (+)-catechin (C), (+)-catechin gallate (CG), (−)-epicatechin (EC), (−)-epicatechin gallate (ECG), (−)-epigallocatechin (EGC), (−)-epigallocatechin gallate (EGCG), (+)-gallocatechin (GC), and (+)-gallocatechin gallate (GCG). Gallic acid (GA), three purine alkaloids of caffeine (CAF), theobromine (TB), and theophylline (TP) were also determined. The contents (mg/kg) were log-transformed to increase the clarity of the figure, and the size of the bubbles represented their contents. The letters in the bubbles represented Tukey's test following one-way ANOVA compared between the five tea cultivars. AB: 'Baiye 1' , FD: 'Fuding Dabaicha' , HJ: 'Huangjinya' , LJ: 'Longjing 43' , YJ: 'Yujinxiang'.
-

Figure 2.
Diversity and composition of phyllosphere microbiome in five tea cultivars. (a) Comparisons of microbial alpha diversity (Shannon index). Letters indicated the significant posthoc test results between treatments (Tukey's test). (b) Principal coordinates analysis (PCoA) of microbial beta diversity. Bray-Curtis dissimilarity was calculated based on the microbial composition at the genus level. (c) Genus composition of microbial communities (relative abundance > 1%). AB: 'Baiye 1' , FD: 'Fuding Dabaicha' , HJ: 'Huangjinya' , LJ: 'Longjing 43' , YJ: 'Yujinxiang'.
-

Figure 3.
Composition of microbial functional genes. (a) Functional pathways of microbial genes. Microbial genes related to nitrogen, sulfur, phosphorus, and methane cycling were analyzed. Genes were further aggregated into functional pathways, and relative abundance was shown. (b) Comparison of gene pathways between tea cultivars. The pathways with relative abundance greater than 1% were compared by one-way ANOVA and Tukey's test, and the significant comparisons were shown. Letters indicated the significant difference between tea cultivars. AB: 'Baiye 1, FD: 'Fuding Dabaicha' , HJ: 'Huangjinya' , LJ: 'Longjing 43' , YJ: 'Yujinxiang'.
-

Figure 4.
Correlations between microbial community, functional genes, amino acids, and 12 secondary metabolites. Four correlation networks were built independently for microbial genera, genes, amino acids, and 12 secondary metabolites. Networks were enhanced to remove the undirected edges. To increase clarity, only the microbial genes with the greatest number of edges were shown, and the names of microbial genera were hidden. The connections between the four networks indicated significant correlations of Mantel tests based on the Bray-Curtis dissimilarity, and insignificant results were not shown.
-
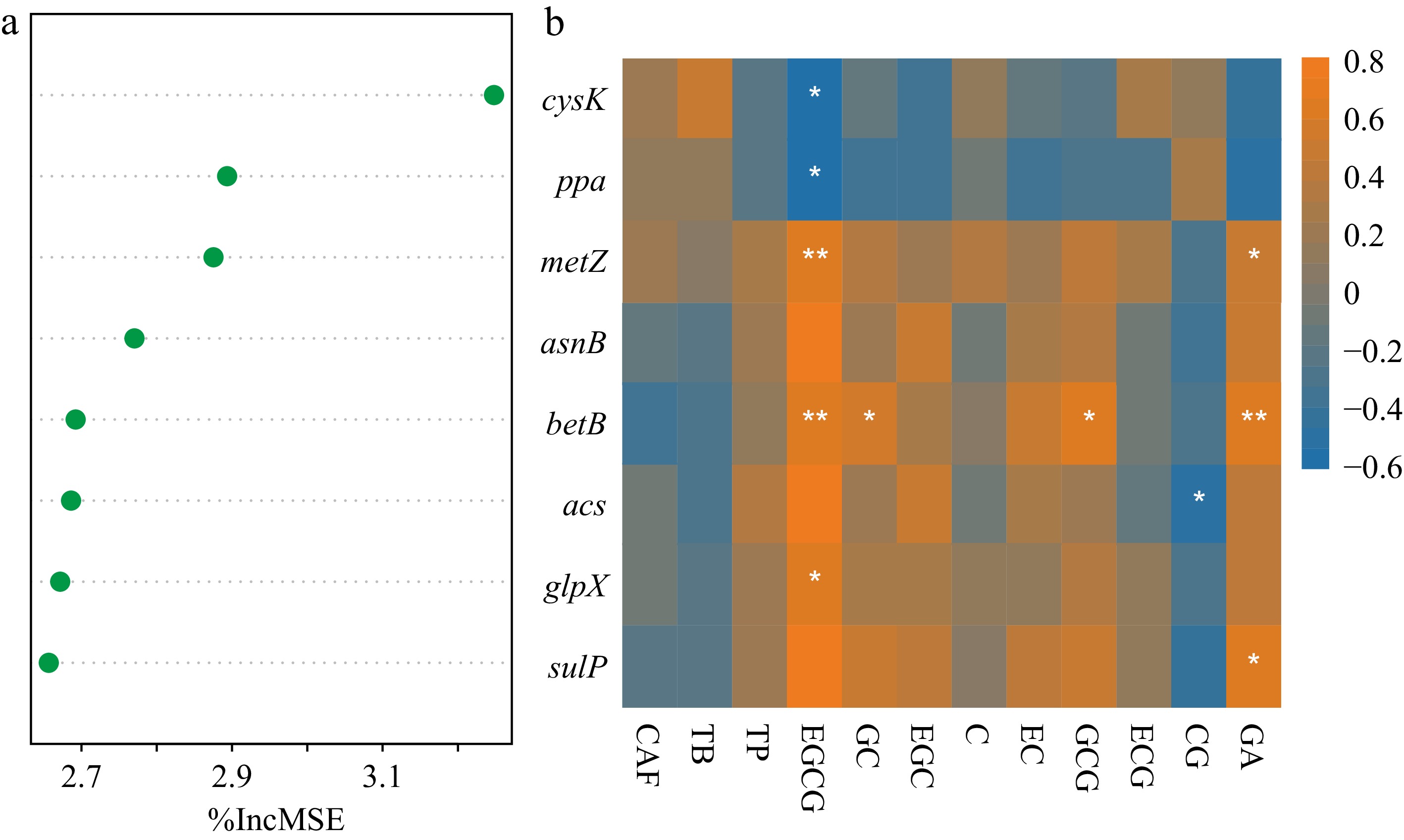
Figure 5.
Featured functional genes in association with 12 leaf secondary metabolites. (a) Eight functional genes identified in the random forest model. The first principal component (PC1) of 12 secondary metabolites was regressed with gene composition. The importance of genes was ranked decreasingly. (b) Correlations between the eight important genes and the examined secondary metabolites. Asterisks indicated significant correlations (p < 0.05). The determined catechins included (+)-catechin (C), (+)-catechin gallate (CG), (−)-epicatechin (EC), (−)-epicatechin gallate (ECG), (−)-epigallocatechin (EGC), (−)-epigallocatechin gallate (EGCG), (+)-gallocatechin (GC), and (+)-gallocatechin gallate (GCG). Gallic acid (GA), three purine alkaloids of caffeine (CAF), theobromine (TB), and theophylline (TP) were also determined.
-
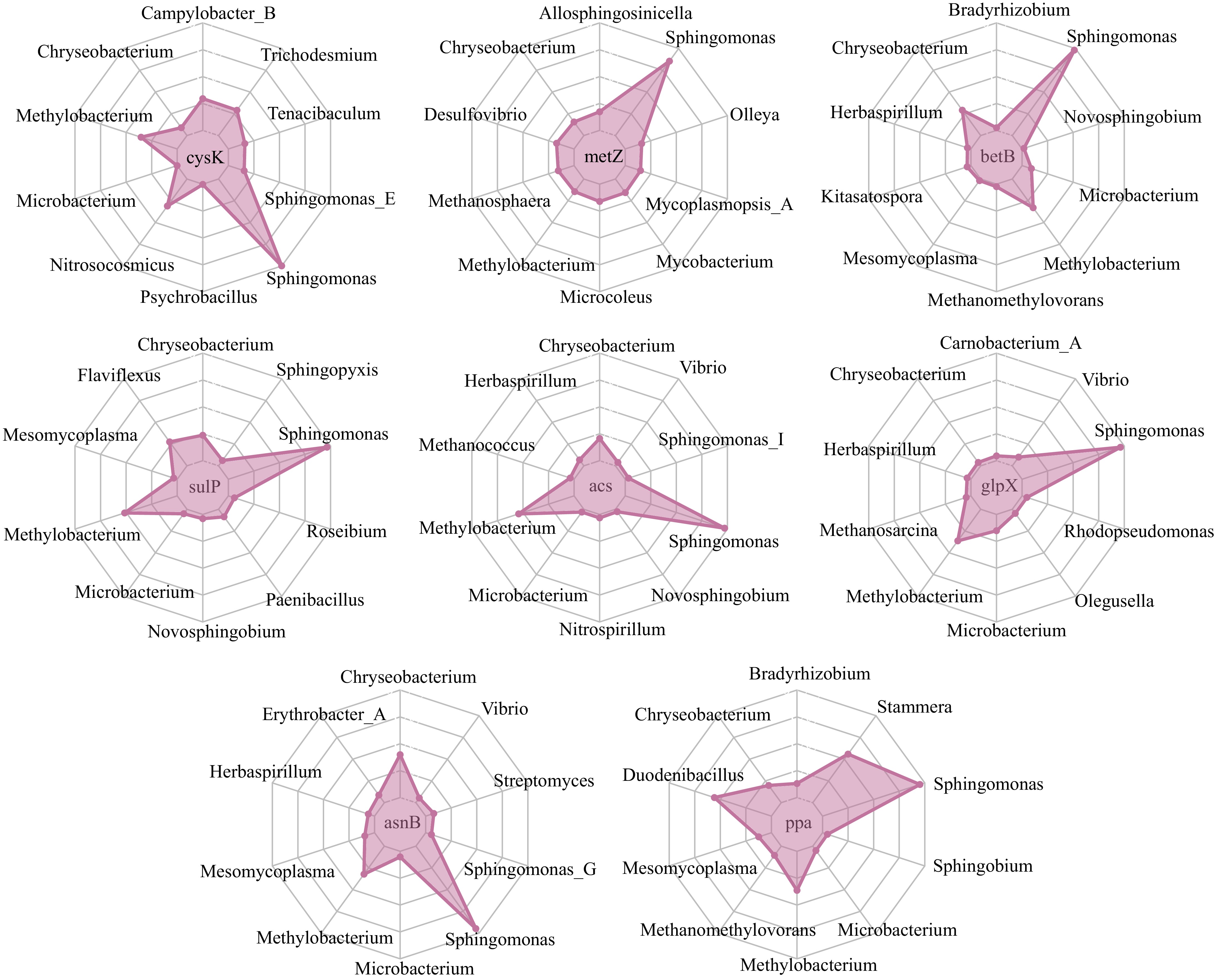
Figure 6.
The taxonomic attribution of microbial functional genes. Eight microbial functional genes were identified by the random forest regressor fitting 12 leaf secondary metabolites on the genera composition of leaf microbiome in five tea cultivars. The contigs containing the gene sequences were then counted and assigned to their belonging genera.
Figures
(6)
Tables
(0)