-
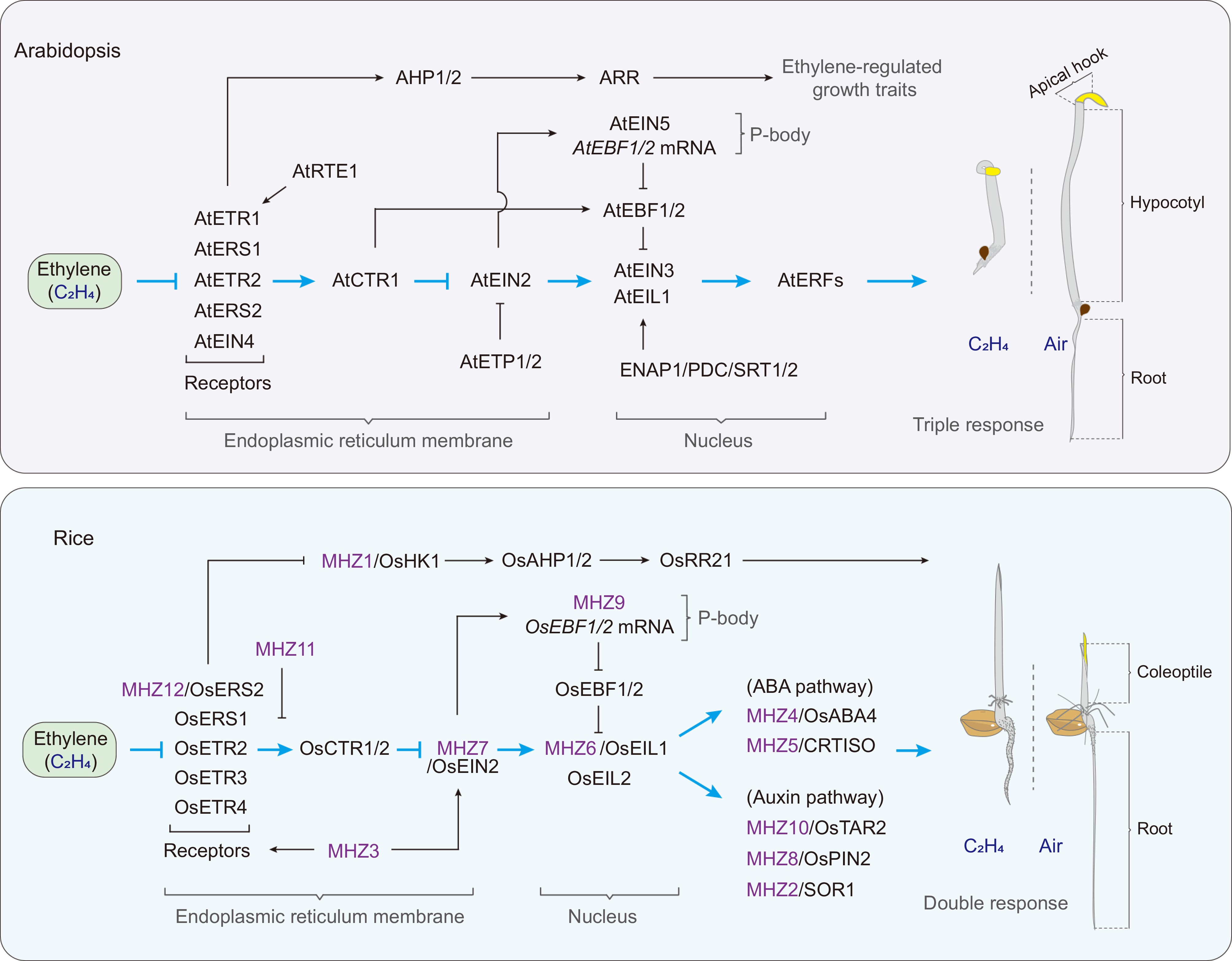
Figure 1.
Model of ethylene signal transduction in Arabidopsis and rice. In Arabidopsis, ethylene receptors, AtCTR1, and AtEIN2 form a complex on the endoplasmic reticulum membrane. Ethylene binding inactivates the receptor/CTR1 complex, relieving repression on AtEIN2. The cleaved EIN2-CEND then follows two pathways: one part enters the P-body, where it inhibits AtEBF1/2 mRNA translation via AtEIN5, while the other translocates to the nucleus to regulate downstream gene expression with AtEIN3/EIL1, ENAP, PDC, and SRT1/2. AtRTE1 and AtETP1/2 act as negative regulators of ethylene signaling, targeting AtETR1 and AtEIN2, respectively. AtCTR1 also can enter the nucleus to repress AtEBF1/2, exerting a positive regulatory role. Also, a non-canonical pathway exists in which AtETR1 signals to histidine-containing AHP, which subsequently activates ARR to modulate ethylene-mediated growth traits. The ethylene receptor-OsCTR1/2-OsEIN2-OsEIL1/2 pathway in rice is highly conserved compared to Arabidopsis. There also exists a distinct MHZ1 pathway independent of OsEIN2. Furthermore, several new regulatory components have been identified: MHZ11 regulates signaling by inhibiting the activation of the ethylene receptor/OsCTR2 complex, while MHZ3 not only stabilizes OsEIN2 but also enhances the activity of the ethylene receptor/OsCTR2 complex. MHZ9 is involved in the translation repression of OsEBF1/2 mRNA in the P-body. The ABA and auxin signaling pathways also interact with the ethylene signaling pathway to co-regulate double responses in rice. The light blue arrows represent the canonical ethylene signaling pathway, while the black arrows indicate non-canonical ethylene signaling pathways or regulation of the components of the canonical ethylene signaling pathway. The 'T' blunt ends indicate negative regulation.
-
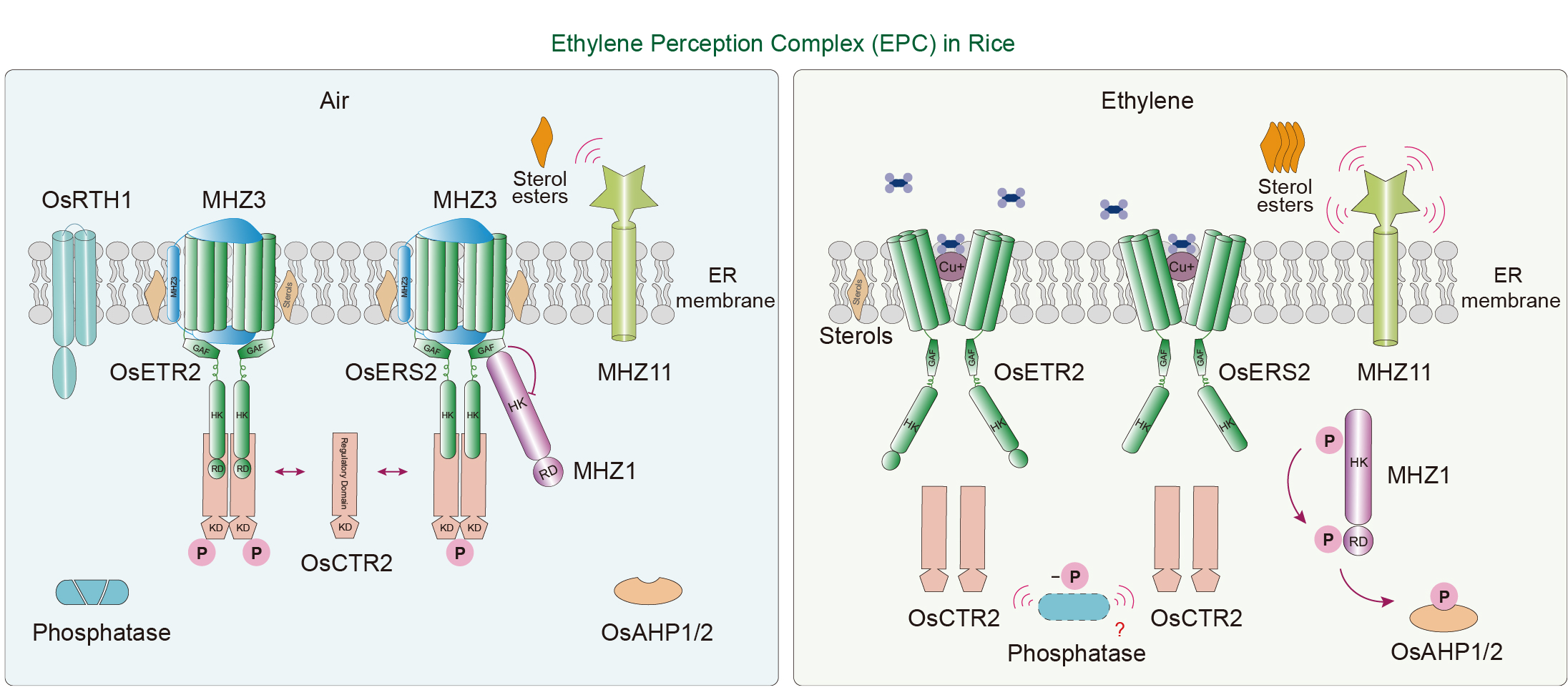
Figure 2.
Ethylene perception complex (EPC) in rice. In the absence of ethylene (left), an EPC is formed with ethylene receptors at its core, preventing signaling activation. MHZ3 interacts with OsETR2 and OsERS2 to stabilize OsCTR2 on the ER membrane, keeping it autophosphorylated and inhibiting the canonical pathway. The GAF domains of ethylene receptors also bind histidine kinase MHZ1, suppressing its activity and blocking the phospho-relay pathway. Upon ethylene perception by the receptors (right), the EPC dissociates, triggering the activation of ethylene signaling. Ethylene binding weakens the interaction between the receptors and MHZ3, leading to the dissociation of OsCTR2. An unknown specific phosphatase may dephosphorylate OsCTR2, inactivating its inhibition of the canonical pathway. Meanwhile, MHZ1 dissociates from the EPC, activating the MHZ1-mediated phospho-relay pathway, transferring phosphate groups to OsAHP1/2, and activating the non-canonical ethylene signaling pathway. Under the influence of ethylene, MHZ11 converts membrane sterols into sterol esters, potentially increasing membrane fluidity and promoting the dissociation of the EPC. The arrows indicate biological processes. The 'T' blunt end indicates negative regulation.
-
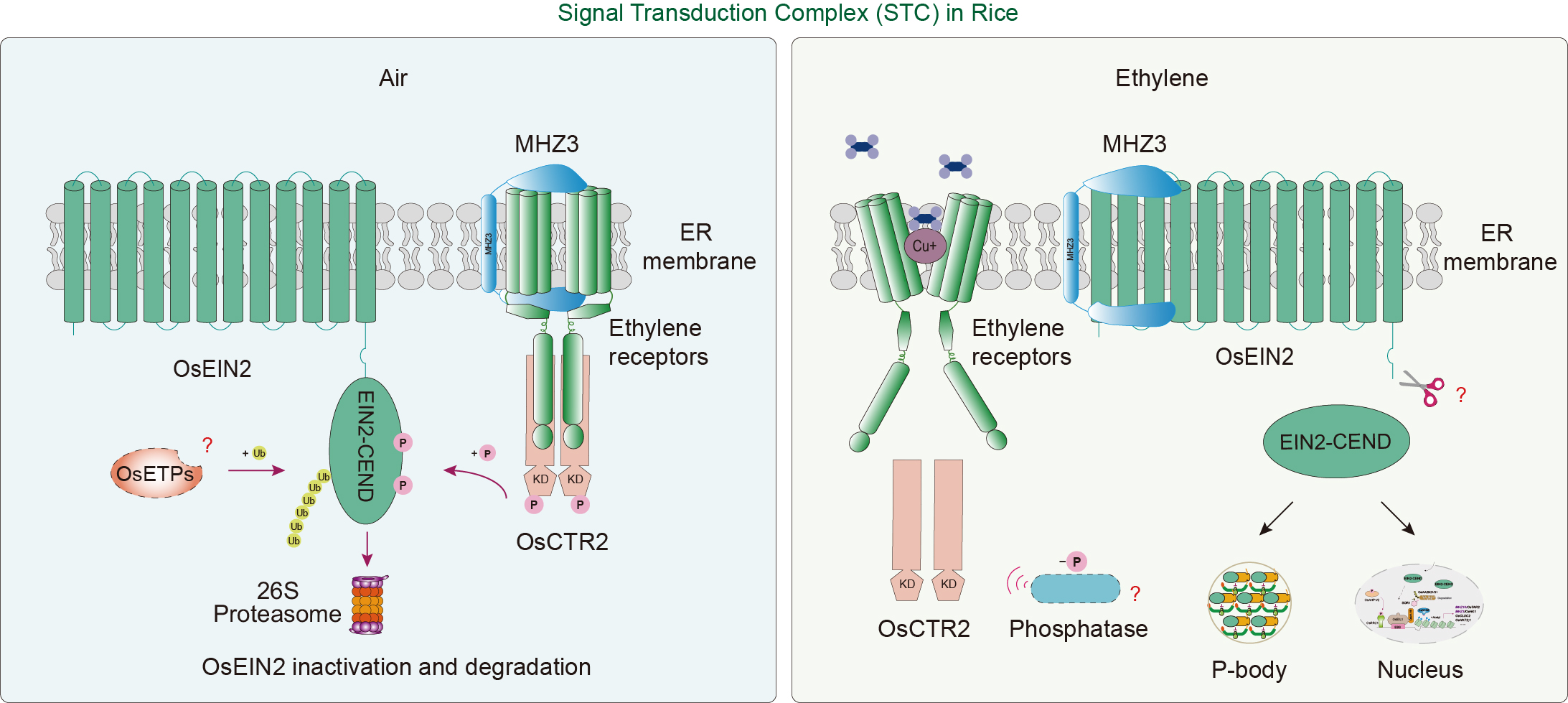
Figure 3.
Signal transduction complex (STC) in rice ethylene signaling. In the absence of ethylene (left), the STC centered on OsEIN2 is unstable. OsEIN2 undergoes phosphorylation by the serine/threonine kinase OsCTR2 and ubiquitination by potential F-box protein OsETPs. These modifications lead to the inactivation and degradation of OsEIN2 via the 26S proteasome, thereby preventing OsEIN2-mediated ethylene signaling. Upon ethylene binding (right), OsCTR2 loses activity and is unable to phosphorylate OsEIN2. Meanwhile, the membrane protein MHZ3 binds to OsEIN2, inhibiting its ubiquitination and subsequent degradation. The stabilized OsEIN2 is cleaved to produce an active C-terminal fragment, which is partially directed to the P-body and partially translocated to the nucleus, thereby completing ethylene signal transduction. Red arrows indicate biological processes, while the black arrows indicate subcellular translocation.
-
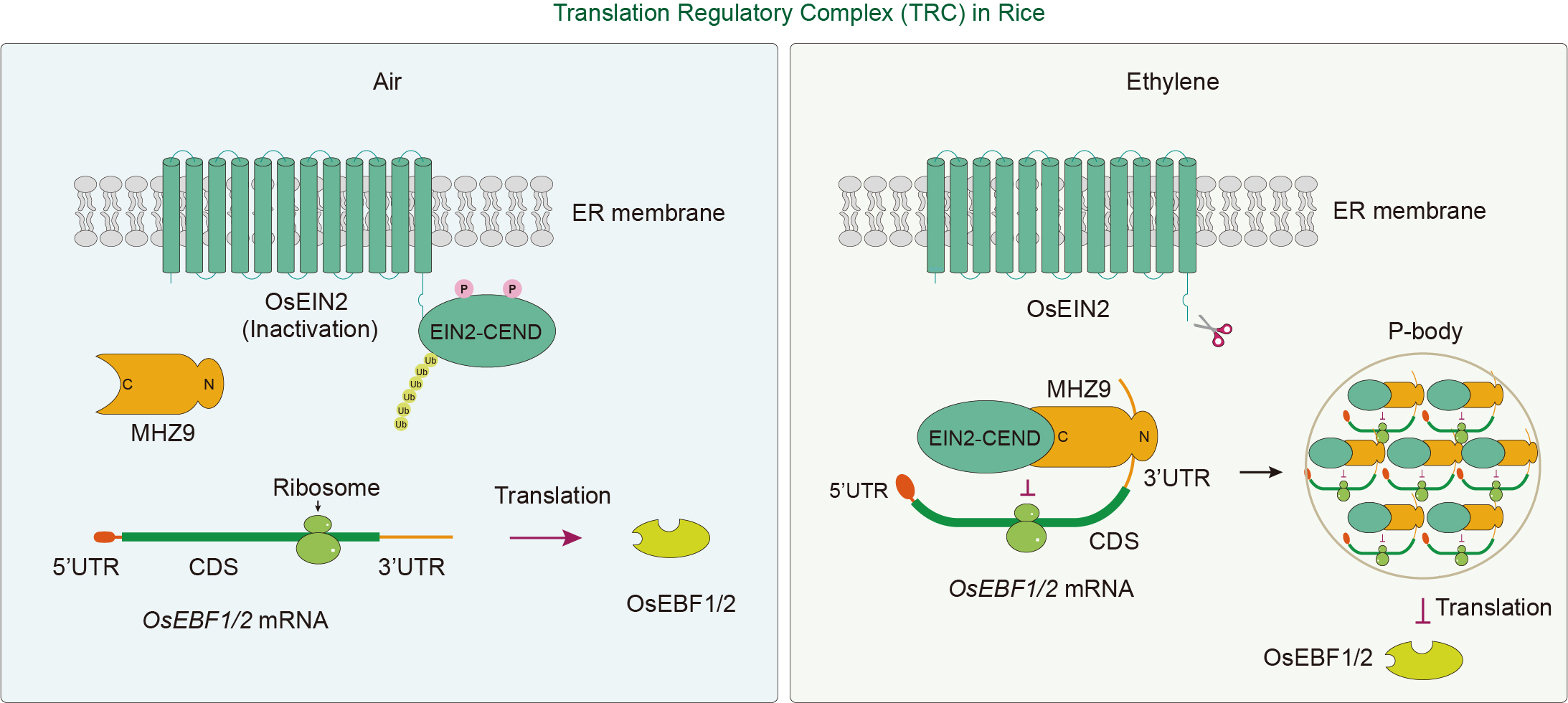
Figure 4.
Translation regulatory complex (TRC) in rice ethylene signaling. In the processing body (P-body), the C-terminal of OsEIN2 (EIN2-CEND), along with MHZ9, OsEBF1/2 mRNA, and other components, forms the TRC, which is involved in the translational inhibition of the ethylene signaling negative regulators, OsEBF1/2 proteins. In the absence of ethylene (left), MHZ9 binds OsEBF1/2 mRNA with low affinity, allowing normal translation and accumulation of the negative regulators OsEBF1 and OsEBF2, which deactivates the ethylene response. In the presence of ethylene (right), the C-terminal region of MHZ9 interacts with OsEIN2-CEND, facilitating the binding of its N-terminal region to the 3' UTR of OsEBF1/2 mRNA, leading to translational repression in the P-body. This prevents OsEBF1/2 accumulation and activates a nuclear ethylene signaling cascade. Dark red arrows indicate biological processes and black arrows indicate subcellular translocation. The 'T' blunt end indicates negative regulation.
-
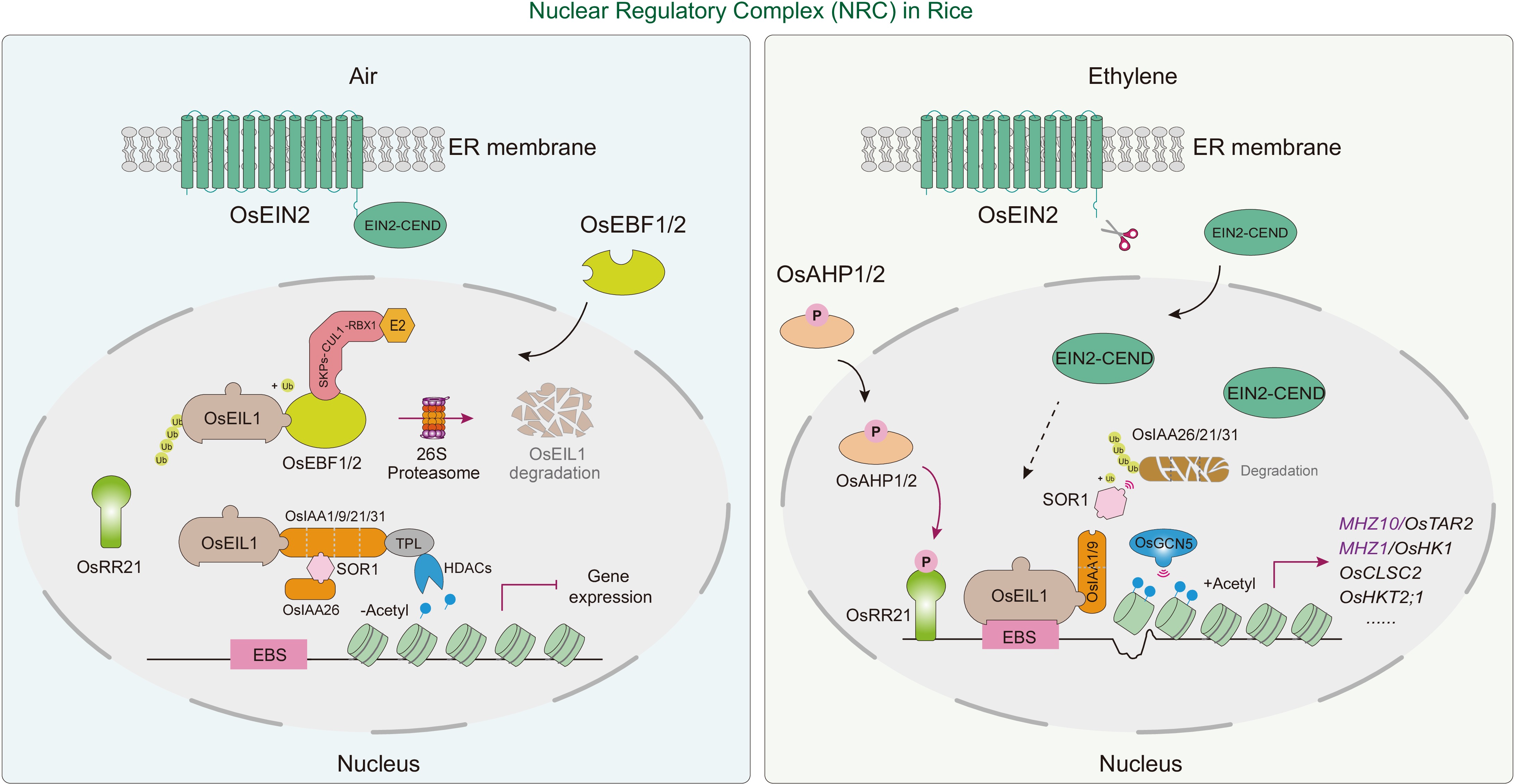
Figure 5.
Nuclear regulatory complex (NRC) in rice ethylene signaling. The core transcription factor OsEIL1 can recruit different interacting components to form a NRC, participating in the regulation of ethylene-responsive genes in rice. In the absence of ethylene (left), the F-box proteins OsEBF1 and OsEBF2 promote the ubiquitination and subsequent degradation of the OsEIL1 transcription factor, thereby inhibiting the activation of ethylene-responsive genes. Low levels of OsEIL1 may interact with auxin repressors OsIAA21 and OsIAA31, repressing downstream genes by recruiting co-repressor TOPLESS (TPL) and histone deacetylases (HDACs), leading to chromatin condensation. Additionally, OsIAA21 and OsIAA31 may attenuate the OsEIL1-OsIAA1/9 complex, maintaining basal biosynthetic processes for normal metabolic functions. In the presence of ethylene (right), OsEIN2-CEND transduces the ethylene signal to the nucleus, where OsIAA1/9 recruits the histone acetyltransferase OsGCN5 to mediate histone acetylation and chromatin decondensation. OsIAA1/9 interacts with OsEIL1, thereby activating the expression of downstream genes. OsEIL1 directly binds to the promoters of MHZ10/OsTAR2, MHZ1/OsHK1, OsCLSC2, OsHKT2;1, and others, playing a role in regulating various biological processes. The response regulator OsRR21, receiving phosphoryl groups from the MHZ1-OsAHP1/2 phospho-relay, activates and amplifies the ethylene signal. Red arrows indicate activation and/or biological process and black arrows indicate subcellular translocation. The 'T' blunt end indicates negative regulation.
-
Arabidopsis Rice Protein Gene No. Phenotype (triple response)
(if unspecified, it is loss-of-function)Ref. Protein Gene NO Phenotype (double response)
(if unspecified, it is loss-of-function)Ref. Ethylene receptors Prokaryote-like histidine kinase AtETR1 AT1G66340 Loss: different degrees of constitutive ethylene response
Gain: ethylene insensitivity[19] OsERS1 Os03g49500 Root ethylene semi-hypersensitivity [52] AtERS1 AT2G40940 [20] MHZ12/
OsERS2Os05g06320 Loss: root ethylene semi-hypersensitivity;
Gain: root-insensitive and coleoptile-semi insensitive ethylene response[50, 52] AtERS2 AT1G04310 [21] OsETR2 Os04g08740 Root ethylene semi-hypersensitivity [4, 52] AtETR2 AT3G23150 [22] OsETR3 Os02g57530 Root ethylene semi-hypersensitivity [51] AtEIN4 AT3G04580 [21] OsETR4 Os07g15540 Unknown [51] Cellular signaling components Serine-threonine protein kinase AtCTR1 AT5G03730 Constitutive triple response [23] OsCTR1 Os09g39320 Root ethylene semi-hypersensitivity [54] OsCTR2 Os02g32610 Root ethylene semi-hypersensitivity [54,55] Nramp-like membrane protein AtEIN2 AT5G03280 Ethylene insensitivity [24] MHZ7/
OsEIN2Os07g06130 Root and coleoptile ethylene insensitivity [49,115] Transcription factor AtEIN3 AT3G20770 Ethylene insensitivity [25] MHZ6/
OsEIL1Os03g20790 Root-insensitive and coleoptile-slightly insensitive ethylene response [53] AtEIL1 AT2G27050 Reduced ethylene sensitivity [25] OsEIL2 Os07g48630 OsEIL2-RNAi:
coleoptile ethylene insensitivity[53] Regulators F-box protein AtEBF1 AT2G25490 ebf1 ebf2: constitutive ethylene response [28−31] OsEBF1 Os06g40360 Hypersensitivity of root and coleoptile [56] AtEBF2 AT5G25350 OsEBF2 Os02g10700 Hypersensitivity of root and coleoptile [56] AtETP1 AT3G18980 amiR-ETP1/ETP2:
constitutive ethylene response[32] Unidentified AtETP2 AT3G18910 Membrane protein AtRTE1 AT2G26070 Enhanced ethylene sensitivity [86] OsRTH1 Os01g51430 Unknown [90] AtMHL1 AT1G75140 mhl1 mhl2: reduced ethylene sensitivity [58] MHZ3 Os06g02480 Root and coleoptile ethylene insensitivity [58] AtMHL2 AT1G19370 Enzyme AtEIN5 AT1G54490 Reduced ethylene sensitivity [116,117] Function not studied Arabidopsis homolog not identified MHZ11 Os05g11950 Root-specific ethylene insensitivity [55] Histidine kinase AtAHK5 AT5G10720 Enhanced ethylene sensitivity [107] MHZ1 Os06g44410 Root-specific ethylene insensitivity [50] RNA-binding protein Arabidopsis homolog not identified MHZ9 Os01g69990 Root-insensitive and coleoptile- semi-insensitive ethylene response [56] Table 1.
Components of Arabidopsis and rice in the ethylene signaling pathway.
Figures
(5)
Tables
(1)