-
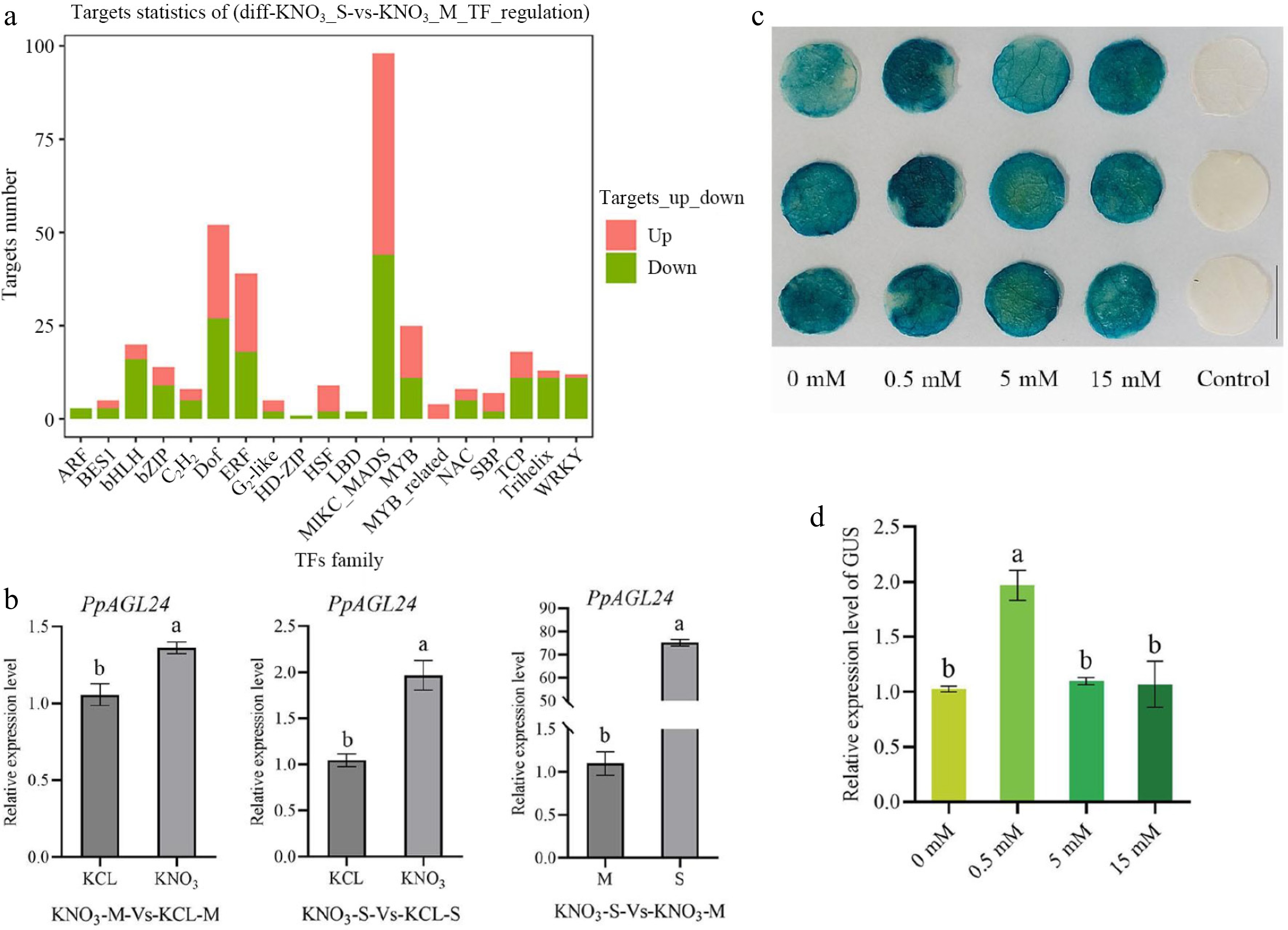
Figure 1.
Transcription factor analysis of differentially expressed transcription factors in 'Shannon-1' (S) and Maotao (M) rootstocks after low nitrogen treatment. (a) TF family with significant DEGs in roots of S and M after 0.1 mM KNO3 treatment. (b) Differential relative expression levels of PpAGL24 in S and M tissues after treatment with 0.1 mM KNO3. Relative transcript levels were normalized to KCl-treated controls (for S) or M baseline (for M), respectively, and expressed as mean ± SD (n = 3). Statistical significance (p < 0.05, Student's t-test) is denoted by distinct lowercase letters above bars. (c) Construct proPpAGL24::GUS reporter infected four-week-old tobacco, performed 0, 0.5, 5, and 15 mM KNO3 100 mL root solution at 0, 24, and 48 h after infection, took 1 cm leaf discs for GUS staining 52 h after infection, and decolorized at 28 °C overnight. The Control group used tobacco leaves injected with a bacterial solution containing empty vectors. (d) Relative expression levels of the GUS gene in tobacco leaf discs were analyzed by RT-qPCR, with transcript levels normalized to the 0 mM KNO3− treated control (GUS). Data are expressed as mean ± SD (n = 3). Distinct lowercase letters indicate statistically significant differences (p < 0.05) determined by one-way ANOVA followed by Duncan's multiple range test.
-
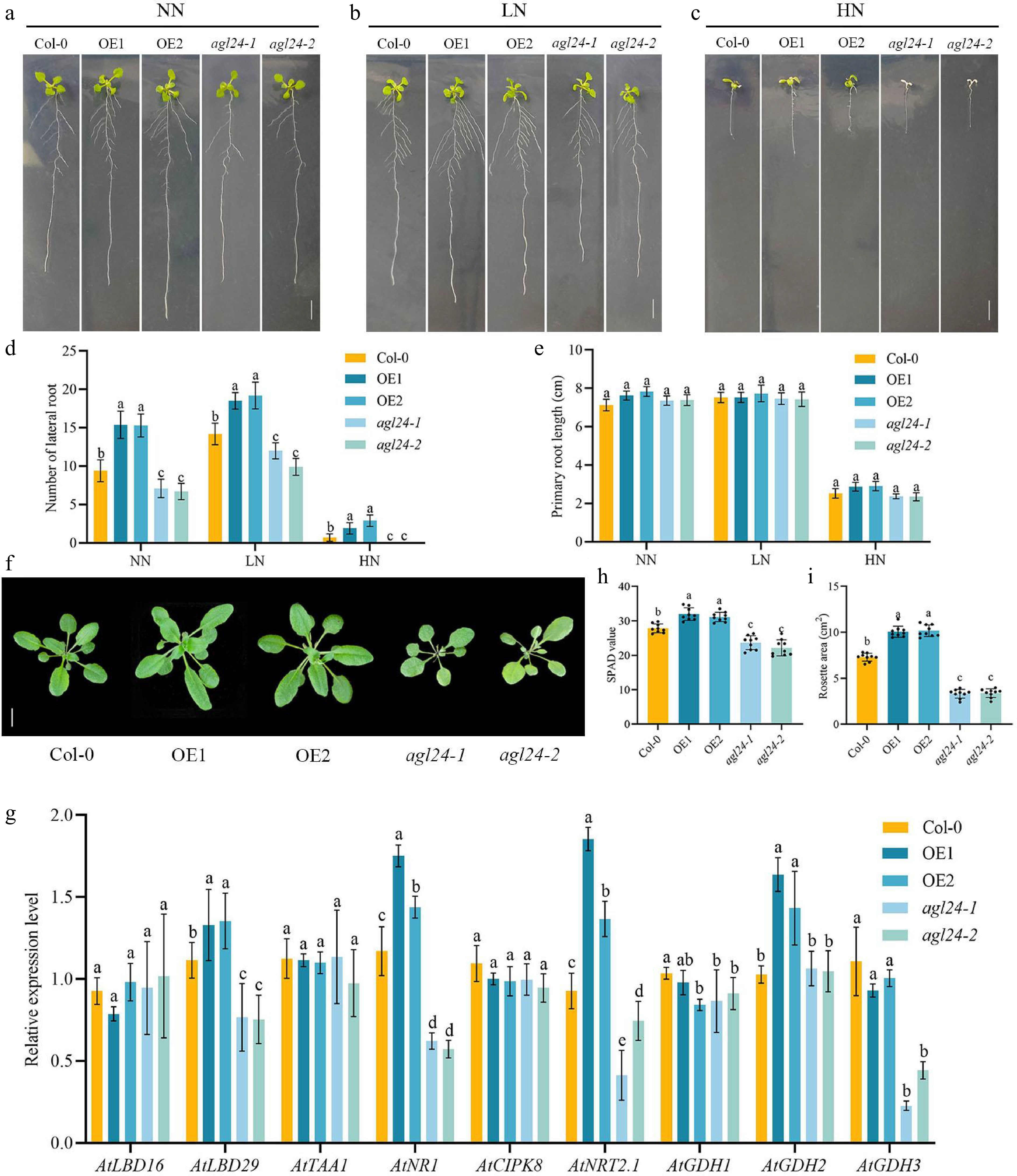
Figure 2.
Analysis of phenotypic characteristics and nitrogen metabolism-related gene expression in Arabidopsis heterologously overexpressing PpAGL24. (a)−(c) Arabidopsis seedlings from overexpressing lines (OE1 and OE2), wild-type Col-0, and AGL24 mutants (agl24-1 and agl24-2) 3 d after germination in MS medium were transplanted to root phenotypes for 7 d in NN, LN, and HN. (d), (e) Number of lateral root length and primary root in Arabidopsis under NN, LN, and HN conditions. (f) Col-0, OE1, OE2, agl24-1, and agl24-2 20-day-old Arabidopsis thaliana shoots. (g) Relative expression levels of genes involved in root development and nitrogen uptake utilization in Col-0, OE1, OE2, agl24-2, and agl24-2 Arabidopsis under NN conditions. (h), (i) SPAD values and rosette leaf area of 20-day-old Arabidopsis. Data represents means ± SD from three independent biological replicates. Significant differences were determined using Duncan's multiple range tests (p < 0.05).
-
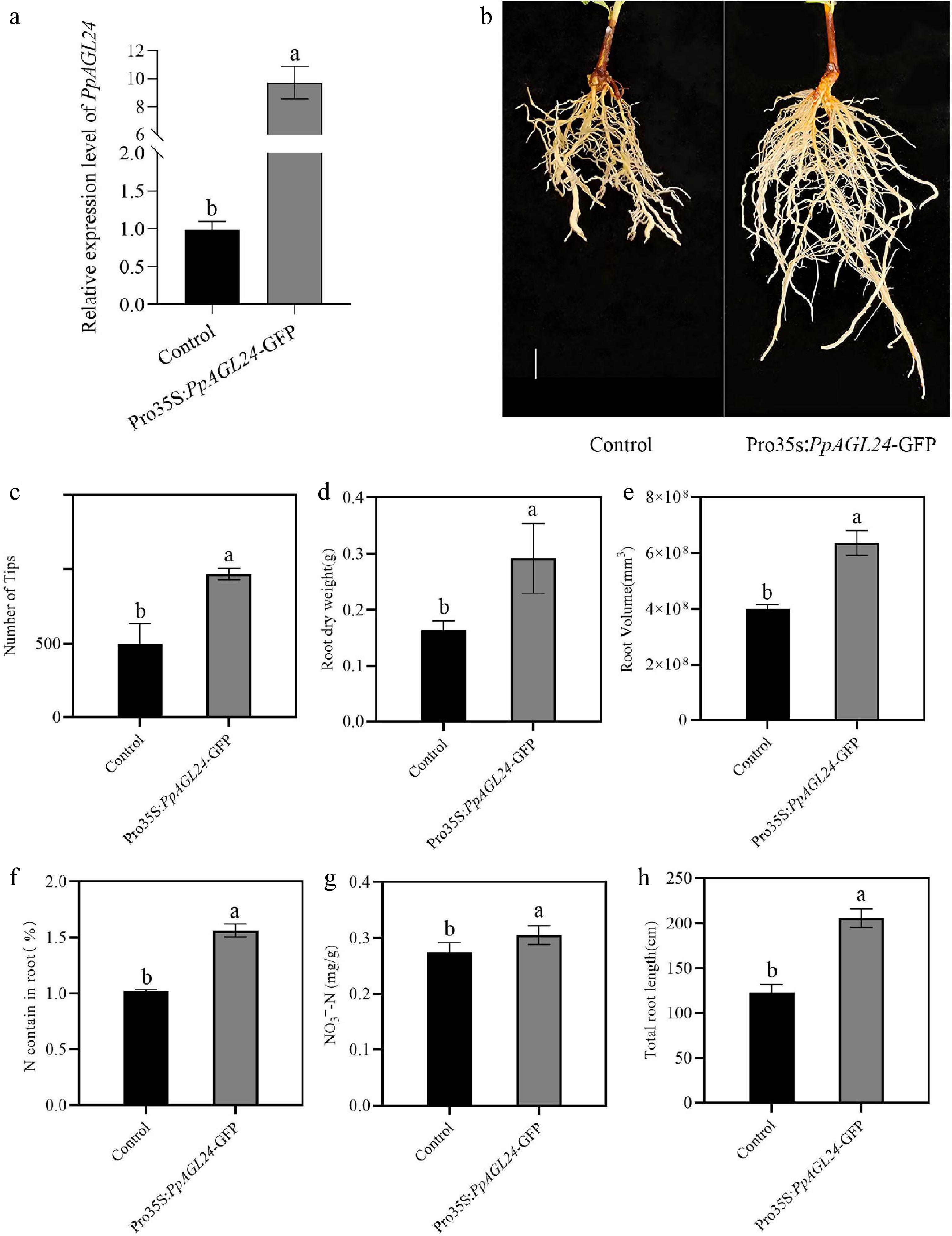
Figure 3.
Overexpression of PpAGL24 promotes peach root development and nitrogen uptake. Pro35s:PpAGL24-GFP represents overexpressed peach roots of PpAGL24, Control represents control transgenic peach roots containing the empty vector. (a) RT-qPCR examination of PpAGL24 transcript levels within control and transgenic root systems. Relative transcript levels were normalised to Control, expressed as means ± SD, n = 3. (b) Phenotypic characterization of regenerated peach roots following Agrobacterium rhizogenes strain K599-mediated transformation. Scale bar = 1 cm. (c)−(i) Number of tips, root dry weight, root volume, N contain in root, NO3−-N contain, total root length. Statistical significance (p < 0.05, Student's t-test) is denoted by distinct lowercase letters above bars.
-
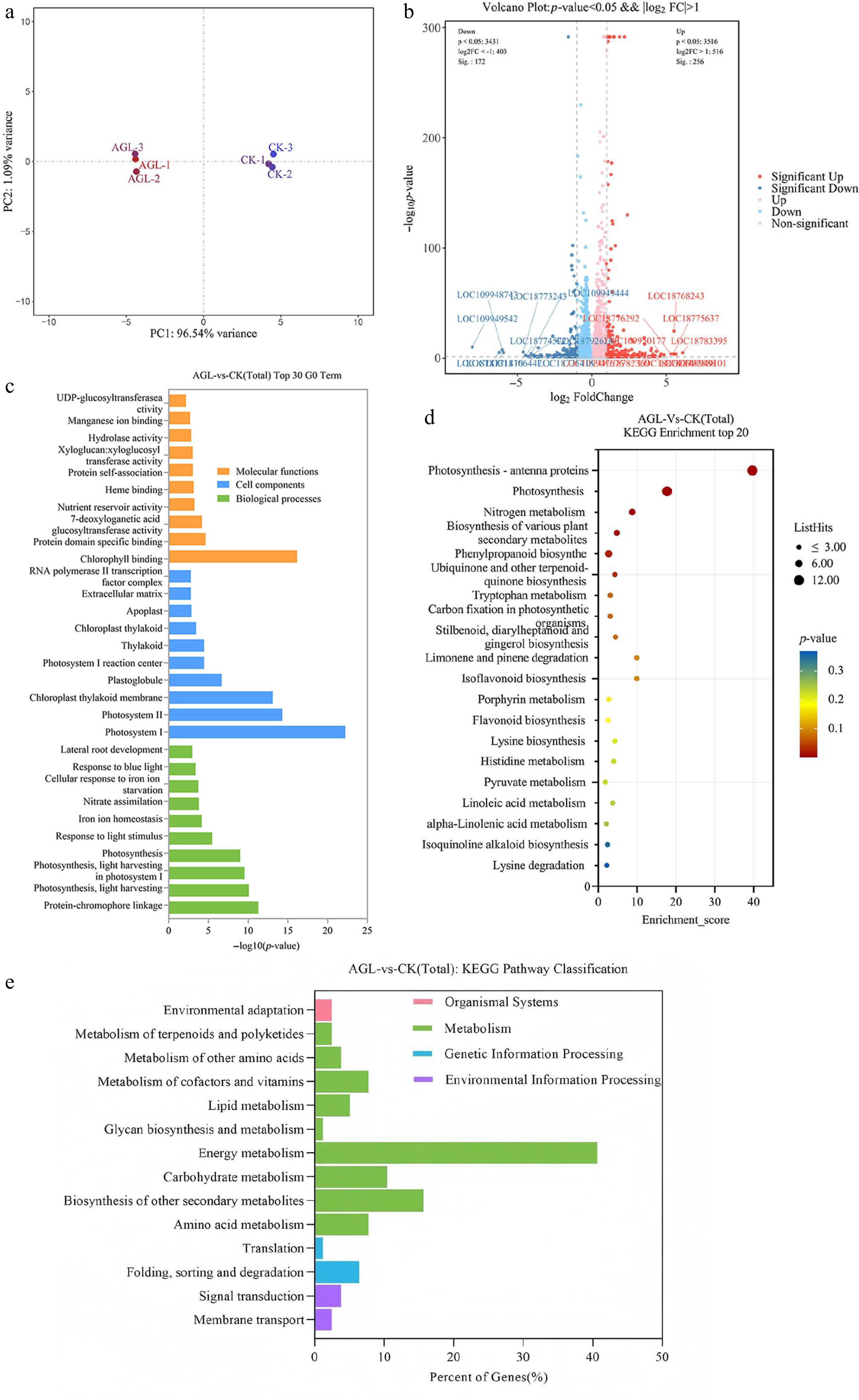
Figure 4.
Comparison of the two transcriptomic libraries. (a) Principal component analysis of the two transcriptomes, AGL-1, AGL-2, and AGL-3: Pro35s: Three biological replicates of peach root overexpressing the PpAGL24-GFP. CK-1, CK-2, and CK-3: three biological replicates of control transgenic peach containing empty vector. (b) Genes differentially expressed in AGL, red scatter are genes upregulated in AGL relative to CK, and blue scatter are downregulated genes. (c) The GO classification of the differentially expressed genes. (d), (e) KEGG pathway enrichment for the differentially expressed genes.
-
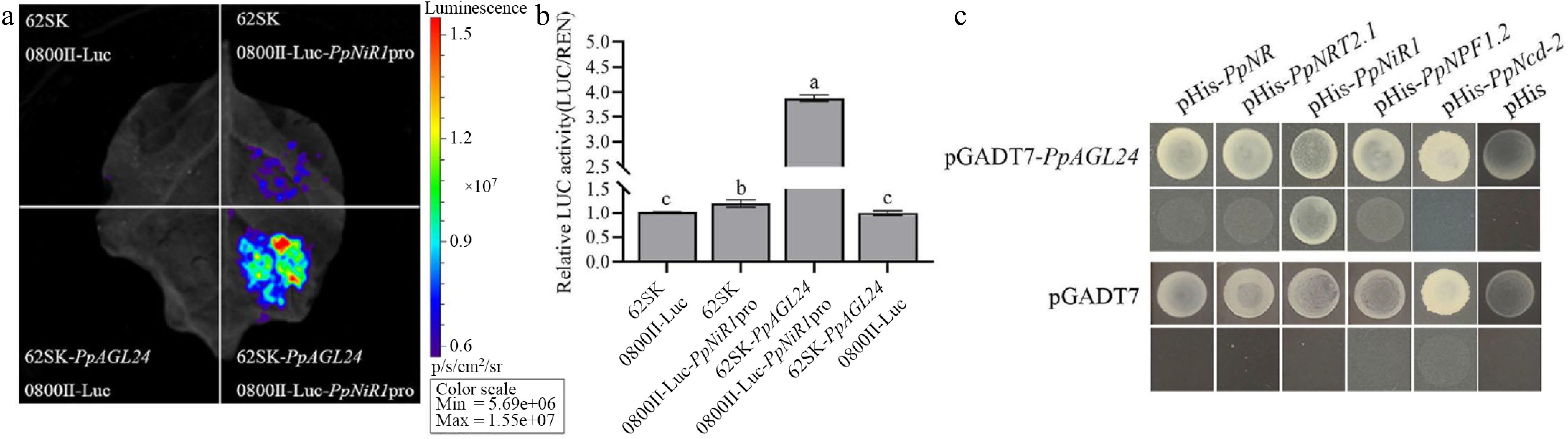
Figure 5.
PpAGL24 is able to activate PpNiR1 transcription. (a) Luciferase imaging analysis. (b) Relative luciferase activity was normalised to LUC/REN for 0800II-Luc empty vector + 62SK empty vector by measuring luciferase activity, expressed as mean ± SD, n = 3. (c) Yeast one-hybrid experiments showed that PpAGL24 was able to bind the promoter of PpNiR1. Different lowercase letters indicate significant differences at p < 0.05 by one-way ANOVA and Duncan (D) multiple comparisons test.
Figures
(5)
Tables
(0)