-
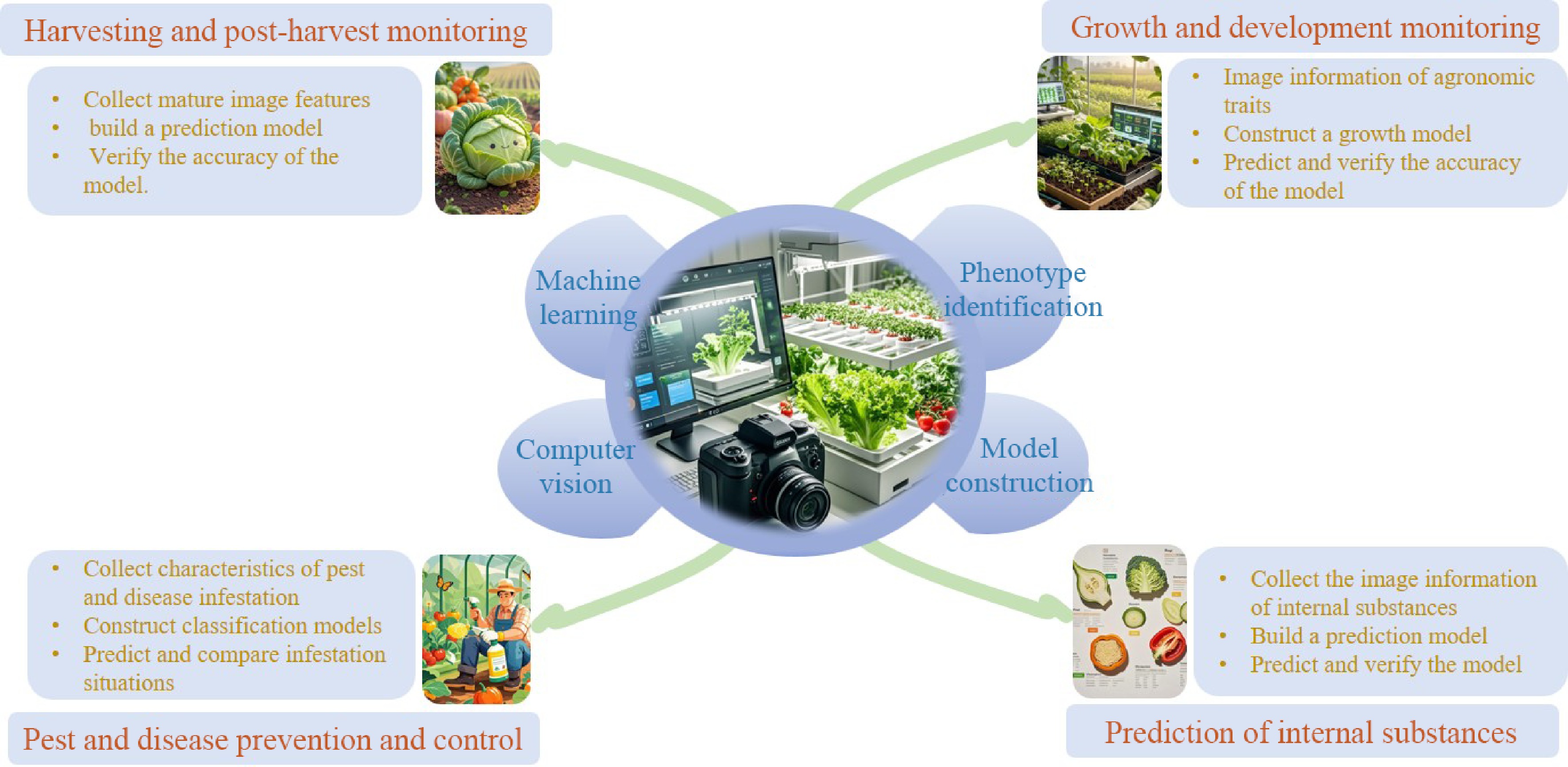
Figure 1.
Research on vegetable phenotypic traits based on computer vision and machine learning.
-
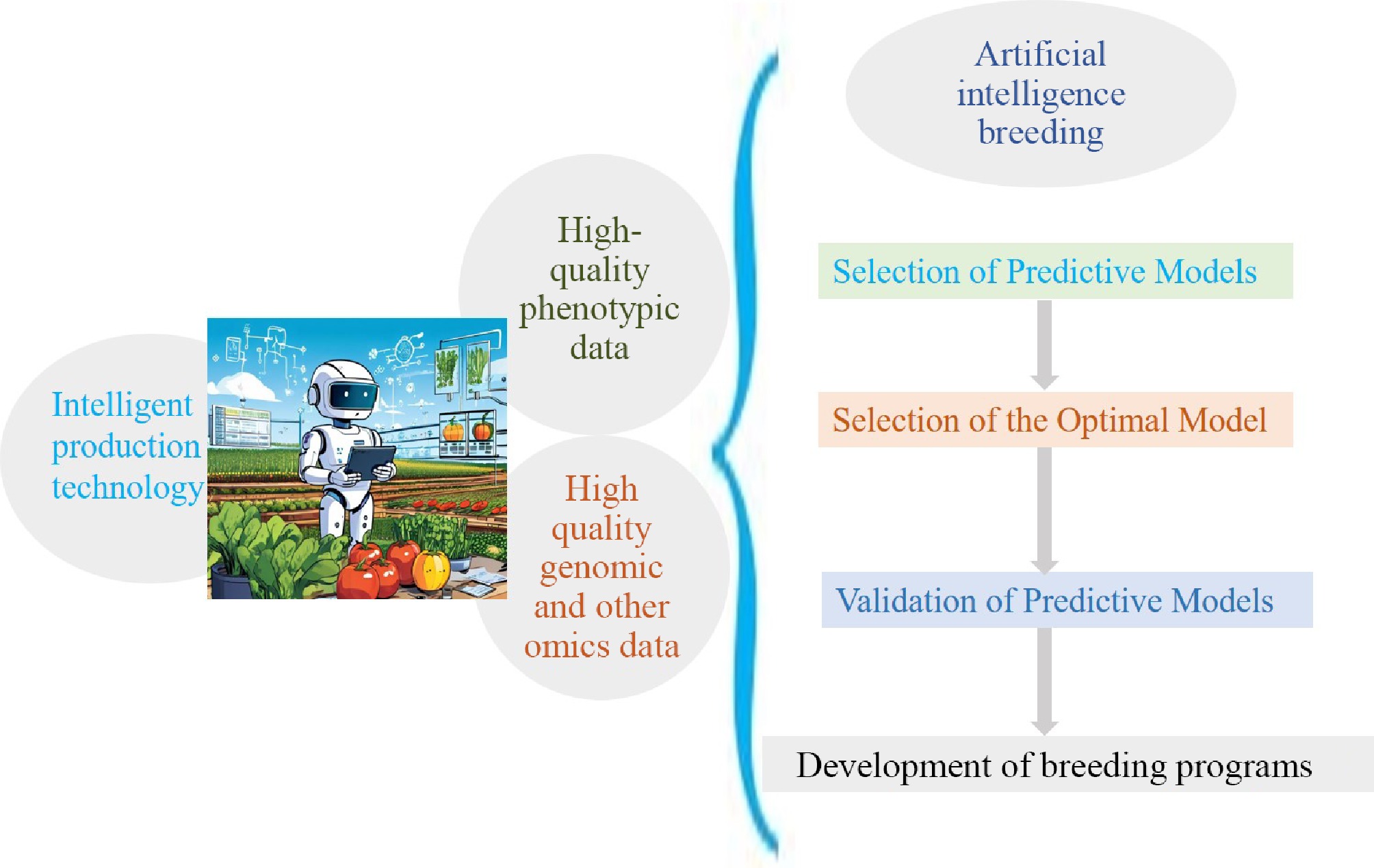
Figure 2.
Schematic diagram illustrating research on the application of AI technology in breeding and related areas.
-
Method Typical algorithms Applications Pros Cons Model complexity Machine learning Linear Reg., Log. Reg., DT, RF, SVM, KNN, etc. Classification, Regression, Clustering, Rec. Systems, etc. Algorithm simplicity, interpretability Limited handling of high-dimensional and non-linear problems Low Deep learning CNN, RNN, LSTM, GAN, etc. Image Recognition, NLP, Speech Recognition, Video Analysis, etc. Automatic feature extraction, effective for high-dimensional and non-linear problems Requires large datasets and computational power, complex models Very high Computer vision Edge Detection, SIFT, SURF, YOLO, SSD, Mask R-CNN, etc. Image Classification, Object Detection, Segmentation, Face Recognition, etc. Specialized for image processing, strong algorithmic focus Relies on manually designed features, limited generalization Medium Image learning Image Enhancement, Repair, Generation (e.g., GAN),
Super-Resolution, etc.Image Enhancement, Repair, Generation, Medical Image Analysis, etc. Focuses on image data, generates high-quality images Requires large datasets, high computational complexity High Table 1.
Comparative analysis of four common AI techniques.
-
Omics field Research topic Research subjects AI methods Application scenarios Key findings Methodological differences Ref. Genomics Novel Whole Genome Selection Method DNNGP Tomato, and others Deep neural network (DNN) Enhancing agronomic trait prediction accuracy DNNGP outperforms on large-scale datasets Higher accuracy but slightly higher computational complexity compared to traditional methods [5] Genomics GS-based tomato fruit quality trait prediction Tomato, pepper, and others Regression models (rrBLUP), classification models (RF, SVC) Optimizing breeding strategies Multi-trait GS model shows stronger prediction for certain traits Random forest and SVM excel in classification tasks, while rrBLUP is more stable in regression tasks [30] Genomics Identification of powdery mildew resistance loci in spinach Spinach GWAS, machine learning (Bayesian B, rrBLUP) Identifying resistance markers Bayesian B model exceeds in resistance prediction High accuracy but longer computation time for Bayesian B model [29] Transcriptomics De novo genome construction of garden mustard Garden mustard Gene prediction (AUGUSTUS), machine learning (logistic regression) Providing genomic resources for breeding and evolutionary studies Identification of 599 potential resistance genes and 459 pre-miRNA encoding sites Logistic regression performs significantly in miRNA site prediction but requires extensive training data [31] Metabolomics Exploration of biological markers for cruciferous vegetable consumption Broccoli, cauliflower, cabbage, radish, mustard, and others Machine learning (random forest classifier) Discovery of food consumption biomarkers Successful identification of metabolites associated with specific cruciferous vegetable consumption High accuracy of random forest classifier in identifying marker metabolites, but risk of overfitting [34] Proteomics Redesign of potato tuber storage proteins Potato Deep learning (alphaFold2, rosetta) Improving functional properties of potato flour Engineered patatin variants significantly enhanced dough viscosity and nutritional value AlphaFold2 provides accurate structure prediction, whereas Rosetta's designed variants are computationally intensive [35] Epigenomics Rational design of plant cis-regulatory sequences Tomato, sunflower, soybean, spinach among 17 plant species Deep learning (transformer) Modeling and designing cis-regulatory variations PhytoExpr model achieves over 85% top-1 accuracy in cross-species mRNA abundance prediction Transformer model excells in multi-task learning but requires substantial computing resources [36] Table 2.
Summary of research progress in vegetable crop multi-omics studies based on AI technology.
Figures
(2)
Tables
(2)