-
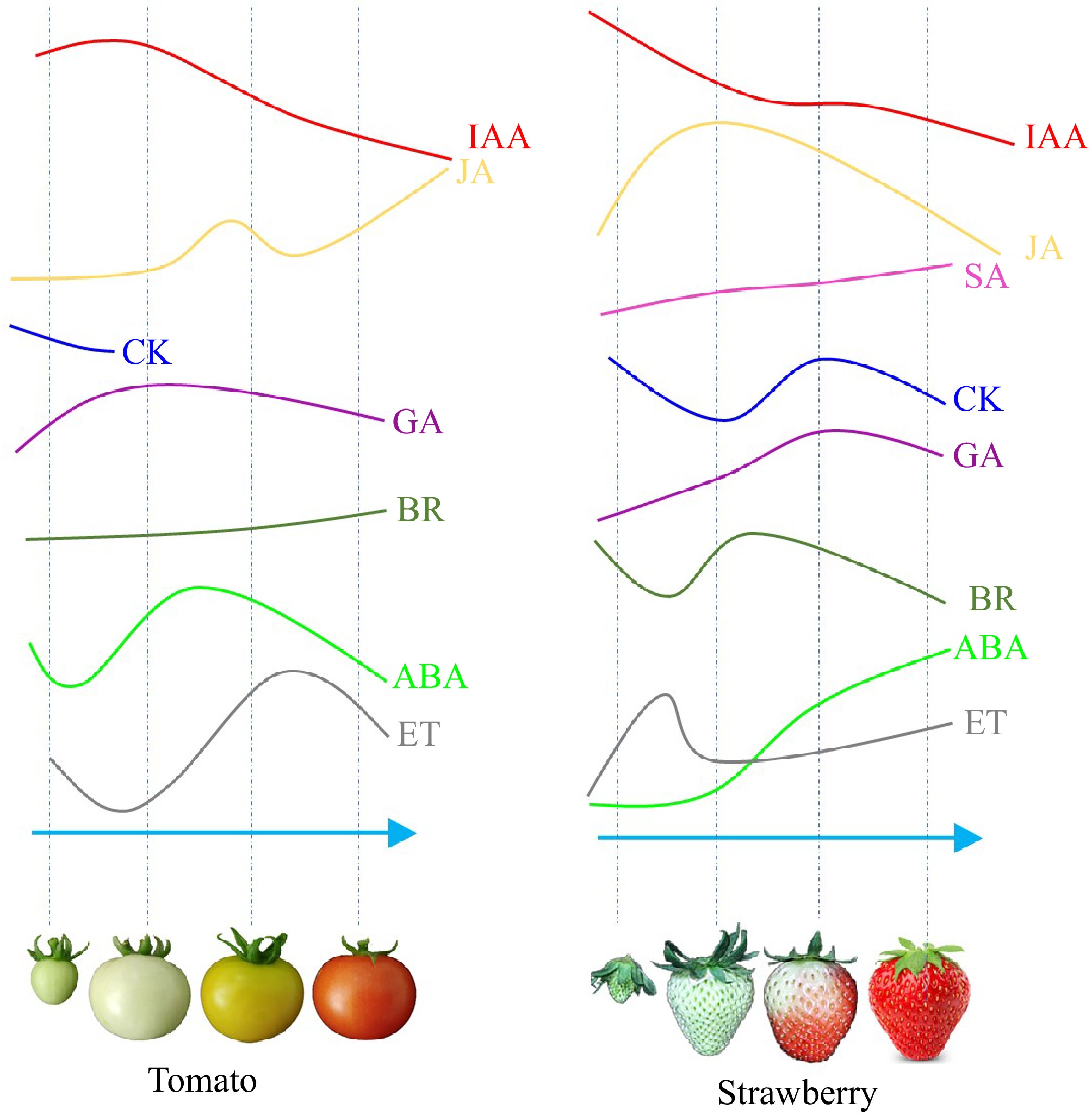
Figure 1.
Schematic diagram of endogenous ET, ABA, IAA, GA, CK, JA, SA, and BR contents in tomato and strawberry during fruit development and ripening. Different coloured lines represent different types of hormones. The horizontal axis represents different developmental stages of tomato and strawberry, while the vertical axis represents the relative contents of the hormones[3,7].
-
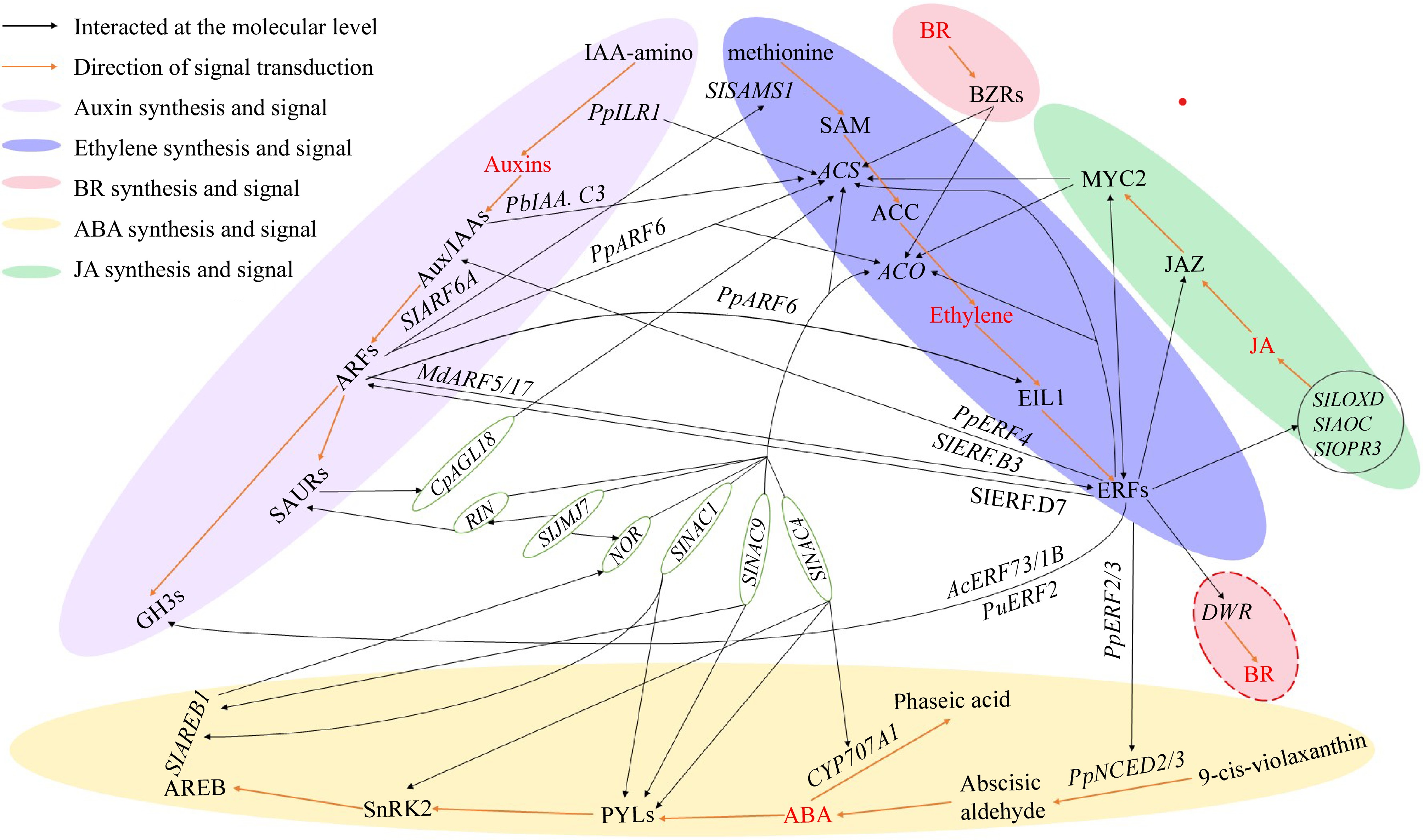
Figure 2.
The molecular crosstalk among ABA, ET, auxin, BR, and JA. The red-colored text represents the five hormones including Auxins, ABA, BR, Ethylene, and JA. The red line represents the hormone synthesis and signal transduction pathway. The black lines represent genes or proteins that have direct interaction relationships which have been confirmed through various experiments including yeast one-hybrid, yeast two-hybrid, EMSA, etc. IAA-amino: indole-3-acetic acid-amino acid; PpILR1: IAA-amino hydrolase gene 1 in peach; Aux/IAAs: auxin/indole-3-acetic acid protein family; SAURs: small Auxin-Up RNAs; GH3s: auxin responsive GH3 gene family; ARFs: auxin response factors; SlSAMS1: S-adenosylmethionine synthetase gene; SAM: S-adenosyl-L-methionine; ACS: 1-aminocyclopropane-1-carboxylate synthase gene; ACC: 1-Aminocyclopropane-1-carboxylic acid; ACO: aminocyclopropanecarboxylate oxidase gene; EIL1: ethylene-insensitive protein; ERFs: ethylene-responsive transcription factors; DWF: steroid 22S-hydroxylase gene, the key for BR biosynthesis; BR: brassinosteroid; MYC2: a basic/helix-loop-helix (bHLH) protein response to JA; JAZ: jasmonate ZIM domain-containing protein; JA: Jasmonic acid; SlLOXD: lipoxygenase gene of tomato; SlAOC: allene oxide cyclase gene of tomato; SlOPR3: 12-oxophytodienoic acid reductase gene of tomato; PpNCED2/3: 9-cis-epoxycarotenoid dioxygenase gene 2/3 in peach; ABA: abscisic acid; CYP707A1: abscisic acid 8'-hydroxylase; PYL: abscisic acid receptor PYR/PYL family; SnRK2: serine/threonine-protein kinase SnRK2: SNF1-related kinase 2; AREB: abscisic acid response element-binding factor; SlNAC1/4/9: NAC transcription factors in tomato; NOR/RIN: two key transcription factor regulating fruit ripening in tomatoes; SlJMJ7: Jumonji C domain-containing protein gene in tomato, an eraser of histone methylation; CpAGL18: a MADS transcription factor in papaya.
-
Nod I Species Nod II Hormone crosstalk Experiment evidence of interaction Ref. SlAREB1 Solanum lycopersicum NOR ABA → NOR Y1H, EMSA, Dual-luciferase Mou et al.[15] SlNAC1 Solanum lycopersicum SlPYL2, SlAREB1 TFs → ABA Y2H, BiFC Yang et al.[19] SlNAC1 Solanum lycopersicum SlACS2, SlACO1 TFs → ET Y1H Ma et al.[18] SlNAC4 Solanum lycopersicum SlACS2, SlACO1, SlACO6, SlACS8 TFs → ET Y2H, BiFC, EMSA Yang et al.[19] SlNAC4 Solanum lycopersicum SAPK3, SlPYL9, SlCYP707A1 TFs → ABA Y2H, BiFC, Y1H, EMSA SlNAC9 Solanum lycopersicum SlAREB1, SlPYL9 TFs → ABA Y2H, BiFC Yang et al.[19] SlNAC9 Solanum lycopersicum LeACO1, LeACS2, LeACS4 TFs → ET EMSA Kou et al.[17] NOR Solanum lycopersicum SlACS2 TFs → ET EMSA Gao et al.[20] SlZFP2 Solanum lycopersicum NOT, SlAO1, SIT, FLC TFs —| ABA ChIP, EMSA, Weng et al.[11] SlJMJ7 Solanum lycopersicum ACS2, ACS3, ACO4, RIN, NOR, DML3 JMJ —| ET ChIP-seq Ding et al.[28] PpERF3 Pyrus pyrifolia PpNCED2/3 ET → ABA Y1H, Dual-luciferase Wang et al.[33] PpERF2 Pyrus pyrifolia PpNCED2/3 ET —| ABA Y1H, EMSA, Dual-luciferase Wang et al.[34] MaEBF1 Musa acuminata MaABI5 ET — ABA Y2H, BiFC, Co-IP Song et al.[36] SlARF2A Solanum lycopersicum SlASR1 Auxin — ABA Y2H, BiFC Breitel et al.[70] SlARF6A Solanum lycopersicum SAMS1 Auxin —| ET EMSA, Dual-luciferase, ChIP Yuan et al.[44] PpILR1 Pyrus pyrifolia PpACS1 Auxin → ET Y1H, EMSA, Dual-luciferase Wang et al.[41] CpARF2 Carica papaya CpEIL1 Auxin → ET Y2H, BiFC, Co-IP Zhang et al.[47] MdARF5 Malus domestica MdACS3a, MdACS1, MdACO1, MdERF2 Auxin —| ET ChIP-PCR Yue et al.[45] CpAGL18 Carica papaya CpACS1, CpSAUR32 TFs → ABA/Auxin Y1H, EMSA, ChIP-qPCR Cai et al.[64] SlERF.D7 Solanum lycopersicum SlARF2A/B ET → Auxin Y1H, EMSA, Gambhir et al.[43] SlERF.B3 Solanum lycopersicum SlIAA27 ET → Auxin EMSA Liu et al.[54] PpERF4 Pyrus pyrifolia PpIAA1, PpNCED2/3 ET — Auxin/ABA Y2H, BiFC, Y1H, EMSA Wang et al.[57] AcERF73/1B Actinidia chinensis AcGH3.1 ET —| Auxin Y1H, Dual-luciferase Gan et al.[61] PuERF2 Pyrus ussuriensis PuGH3.1 ET —| Auxin Y1H, EMSA Yue et al.[62] RIN Solanum lycopersicum SlSAUR69 ET — Auxin Dual-luciferase Shin et al.[40] MdMYC2 Malus domestica MdACO1, MdACS1, MdERF2/3 JA → ET Y1H, EMSA, ChIP-PCR Li et al.[95] MdERF4 Malus domestica MdJAZ, MdMYC2 ET — JA ChIP-seq, BiFC Hu et al.[99] SlERF.B8 Solanum lycopersicum SlLOXD, SlAOC, SlOPR3 ET → JA ChIP-qPCR, Dual-luciferase Ding et al.[98] MaBZR1/2 Musa acuminata MaACS1, MaACO13, MaACO14 BR —| ET EMSA, Dual-luciferase Guo et al.[111] DkBZR1/2 Diospyros kaki DkACS1, AkACO2 BR —| ET EMSA, Dual-luciferase He et al.[112] SlAP2a Solanum lycopersicum DWF ET —| BR Y1H, EMSA, Dual-luciferase Sang et al.[108] → indicates a positive effect, —| indicates a negative effect, and — indicates other interactions. Table 1.
A summary of hormone crosstalk interaction data.
Figures
(2)
Tables
(1)