-

Figure 1.
(a)−(f) The length distribution of miRNAs in six samples. Three repeated samples venn plots of (g) BZP01, and (h) BZP02. The (i) first bias preference, and (j) each point base bias.
-

Figure 2.
The expression levels of miRNAs in BZP01 and BZP02. (a) Venn plots of BZP01 and BZP02. (b) The number of upregulated and downregulated miRNAs. (c) Expression heatmap of 438 miRNAs between BZP01 and BZP02. (d) Volcano plot of BZP01 vs BZP02.
-

Figure 3.
GO and KEGG enrichment analysis of 604 genes targeted by 29miRNAs. (a) GO enrichment analysis. (b) KEGG pathway annotation. (c) The top 20 KEGG pathway enrichment.
-
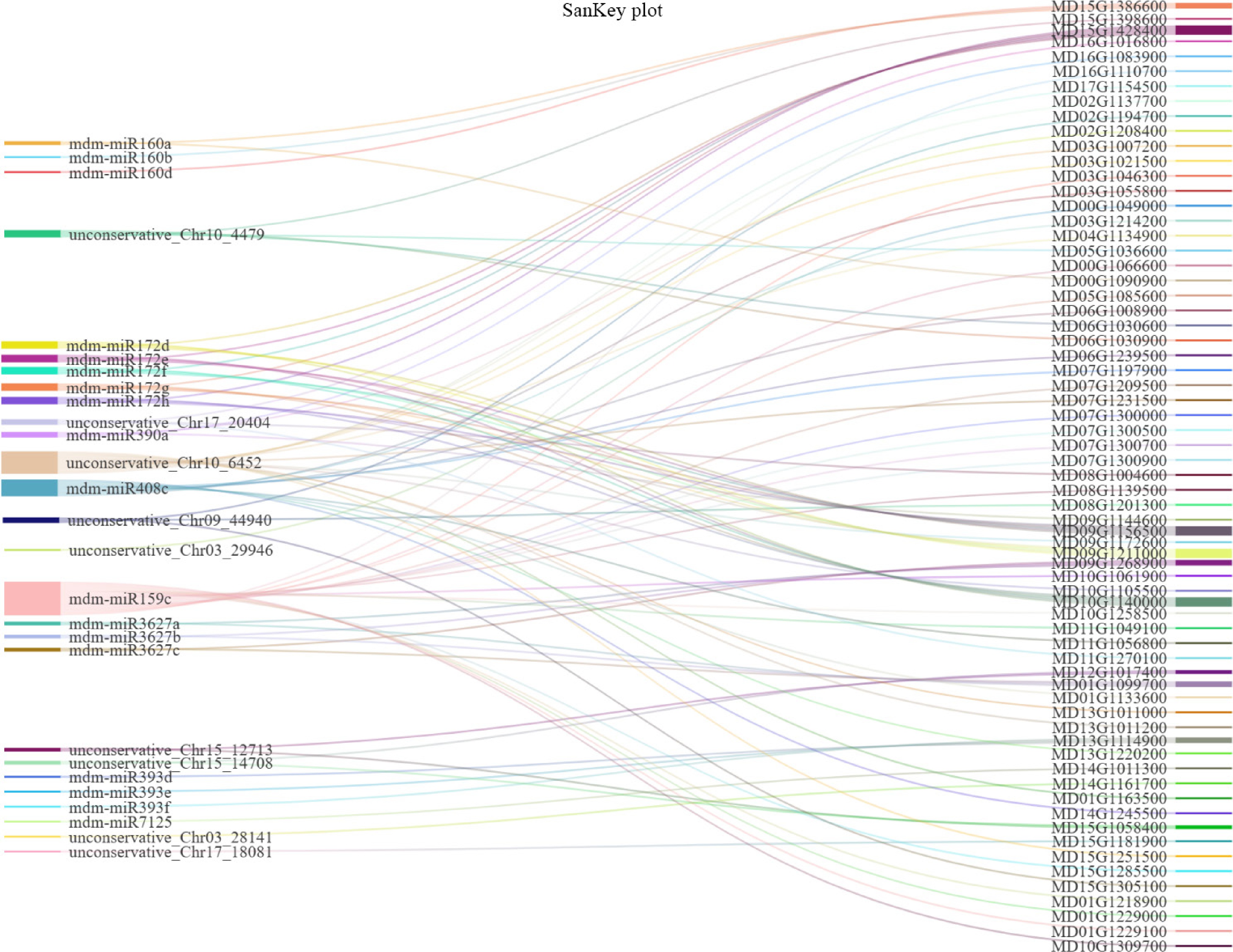
Figure 4.
Sankey diagram of 67 mRNAs and 27 miRNAs. The mRNAs derived from five pathways (highlighted in red boxes, Fig. 3c) associated with physiological changes and VOCs biosynthesis of 'Binzi' peels.
-
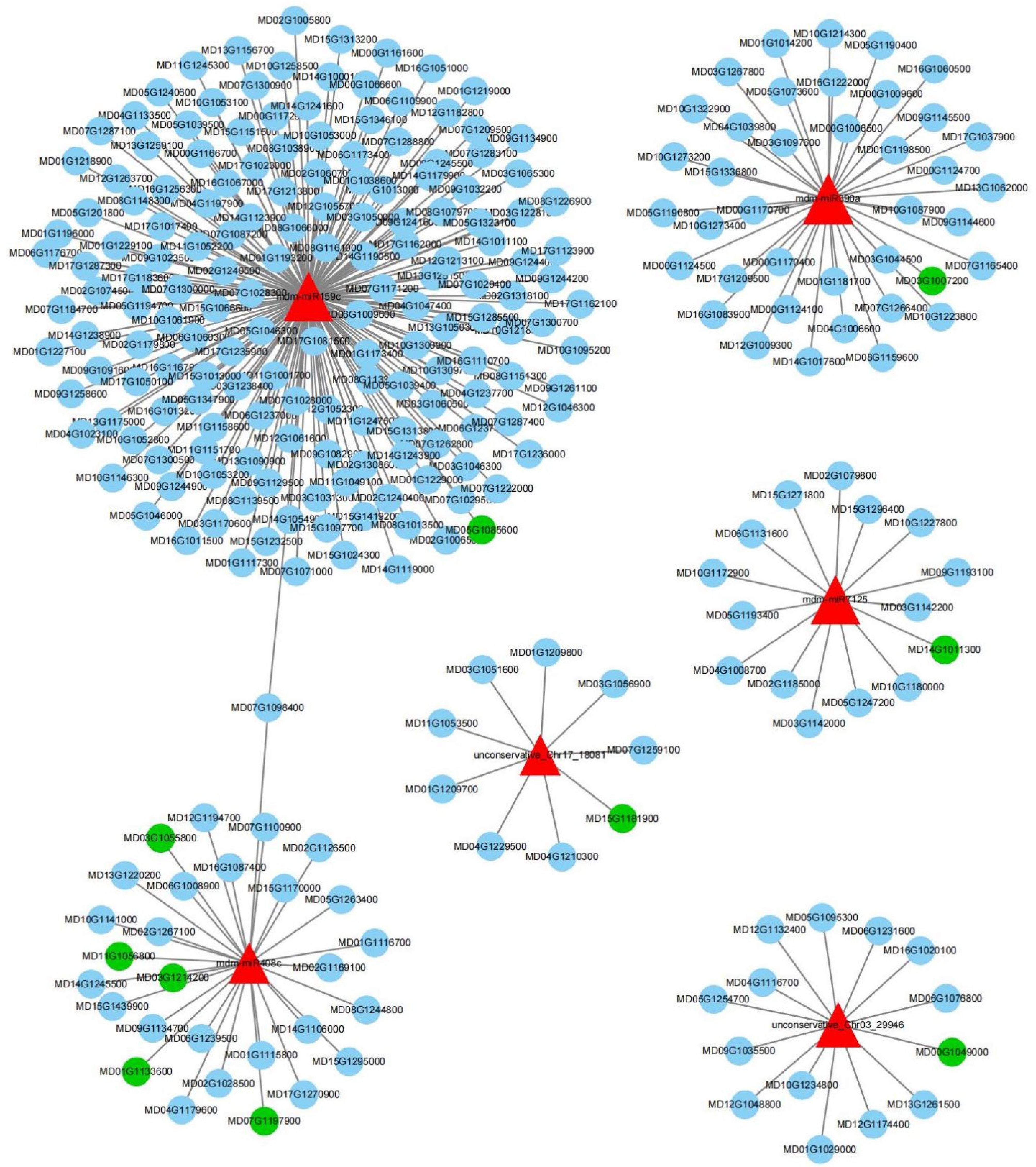
Figure 5.
Network of miRNA and mRNA involved in volatile organic components synthesis. The red triangle represents miRNAs, the blue circles represent targeted genes, and the green circles represent genes related to the pathways of aroma production.
-
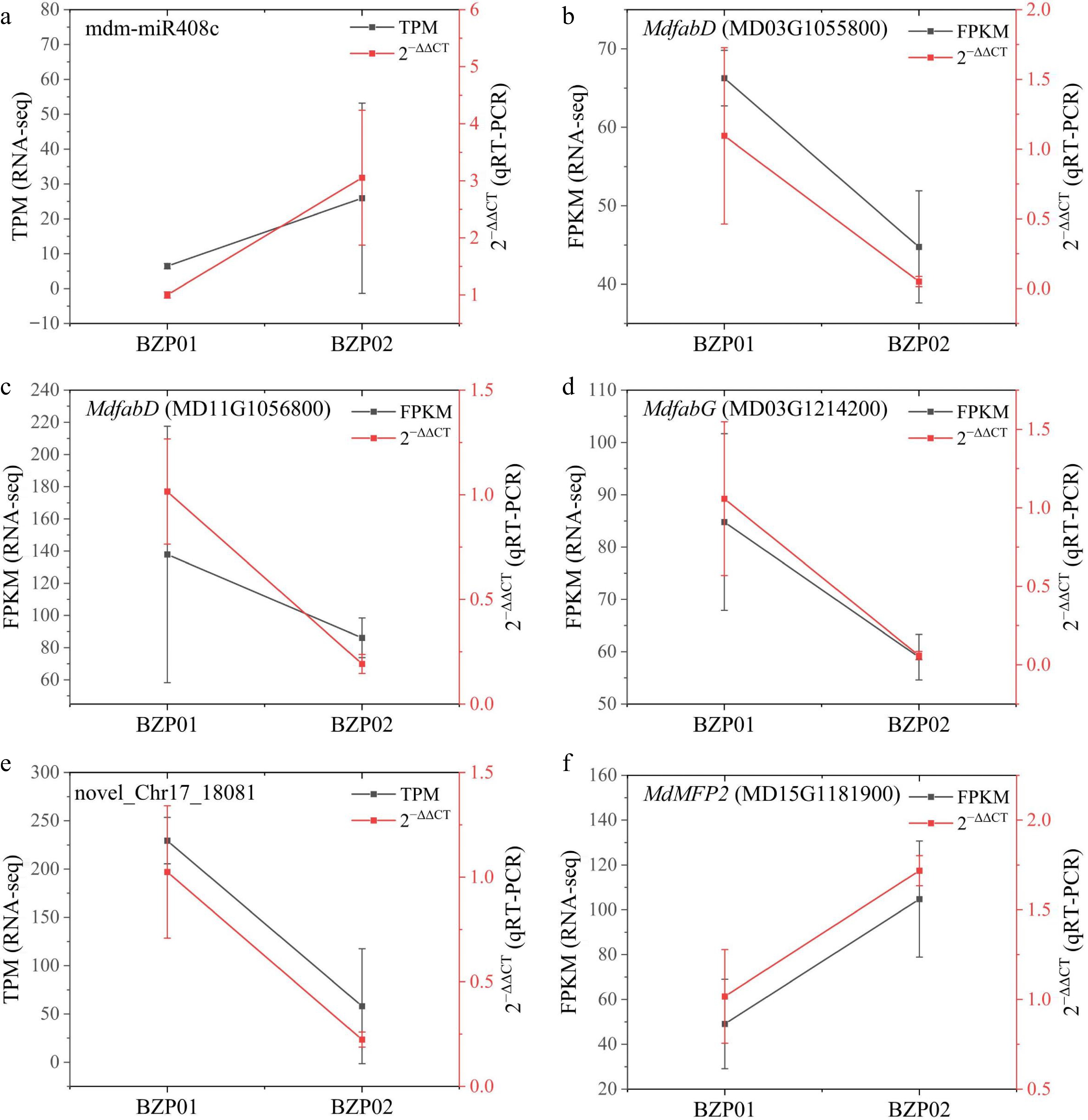
Figure 6.
Transcriptome expression levels and qRT-PCR of miRNAs and target genes. (a) The FPKM and 2−∆∆Ct of mdm-miR408c and its target gene (b) MdfabD (MD03G1055800), (c) MdfabD (MD11G1056800), and (d) MdfabG (MD03G1214200). (e) The FPKM and 2−∆∆Ct of novel_Chr17_18081 and its target gene MdMFP2 (MD15G1181900).
-
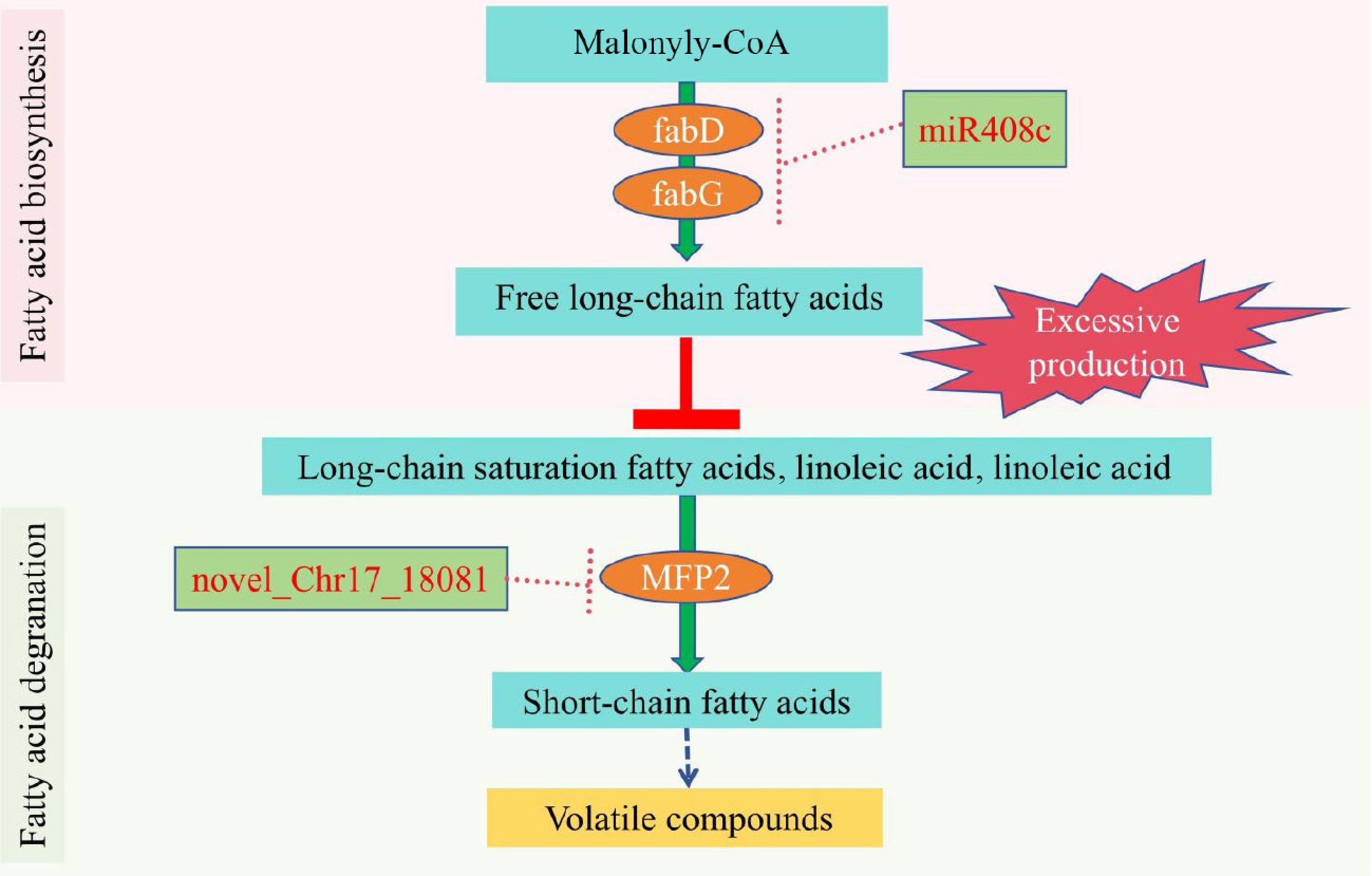
Figure 7.
A working model on the role of miR408c, novel_Chr17_18081, MdfabD, MdfabG, and MdMFP2 in regulating volatile aroma compounds synthesis in 'Binzi' apple. MdfabD: [acyl-carrier-protein] S-malonyltransferase. MdfabG: 3-oxoacyl-[acyl-carrier protein] reductase. MdMFP2: enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase.
-
Compound Metabolism pathway Post-harvest storage period 0 d 5 d 10 d Ethyl butyrate Fatty acid metabolism – 3,439.60 1,727.43 Ethyl caproate Fatty acid metabolism – 1,167.97 782.60 Ethyl 2-methylbutyrate Amino acid metabolism – 1,546.96 383.70 Hexanal Fatty acid metabolism 93.26 171.20 454.31 Ethanol Fatty acid metabolism – 233.95 973.26 Hexyl alcohol Fatty acid metabolism 379.26 1,486.10 1,772.29 (E)-2-Hexen-1-ol Fatty acid metabolism 523.72 – – 6-Methyl-5-hepten-2-one Carotenoid metabolism 228.80 3,674.34 973.51 – not detected. Table 1.
Differential VOCs in 'Binzi' peels during storage (μg/kg)[32].
-
Pathway Pathway ID miRNAs Target genes Target genes name K_id K_id entry Fatty acid metabolism ko01212 mdm-miR408c MD03G1214200 3-Oxoacyl-[acyl-carrier protein] reductase (fabG) K00059 [EC:1.1.1.100] mdm-miR408c MD03G1055800 [acyl-carrier-protein]
S-malonyltransferase (fabD)K00645 [EC:2.3.1.39] mdm-miR408c MD11G1056800 [acyl-carrier-protein]
S-malonyltransferase (fabD)K00645 [EC:2.3.1.39] mdm-miR408c MD07G1197900 Very-long-chain enoyl-CoA reductase (TER) K10258 [EC:1.3.1.93] mdm-miR408c MD01G1133600 Very-long-chain enoyl-CoA reductase (TER) K10258 [EC:1.3.1.93] novel_Chr17_18081 MD15G1181900 Enoyl-CoA hydratase/
3-hydroxyacyl-CoA dehydrogenase (MFP2)K10527 [EC:4.2.1.17, 1.1.1.35, 1.1.1.211] Biosynthesis of unsaturated fatty acids ko01040 mdm-miR408c MD07G1197900 Very-long-chain enoyl-CoA reductase (TER) K10258 [EC:1.3.1.93] mdm-miR408c MD01G1133600 Very-long-chain enoyl-CoA reductase (TER) K10258 [EC:1.3.1.93] Fatty acid biosynthesis ko00061 mdm-miR408c MD03G1214200 3-Oxoacyl-[acyl-carrier protein] reductase (fabG) K00059 [EC:1.1.1.100] mdm-miR408c MD03G1055800 [acyl-carrier-protein]
S-malonyltransferase (fabD)K00645 [EC:2.3.1.39] mdm-miR408c MD11G1056800 [acyl-carrier-protein]
S-malonyltransferase (fabD)K00645 [[EC:2.3.1.39] Fatty acid degradation ko00071 novel_Chr17_18081 MD15G1181900 Enoyl-CoA hydratase/
3-hydroxyacyl-CoA dehydrogenase (MFP2)K10527 [EC:4.2.1.17 1.1.1.35 1.1.1.211] alpha-Linolenic acid metabolism ko00592 novel_Chr17_18081 MD15G1181900 Enoyl-CoA hydratase/
3-hydroxyacyl-CoA dehydrogenase (MFP2)K10527 [EC:4.2.1.17 1.1.1.35 1.1.1.211] Biosynthesis of amino acids ko01230 mdm-miR7125 MD14G1011300 Cysteine synthase (cysK) K01738 [EC:2.5.1.47] Carotenoid biosynthesis ko00906 mdm-miR390a MD03G1007200 15-Cis-phytoene synthase (crtB) K02291 [EC:2.5.1.32] novel_Chr03_29946 MD00G1049000 Lycopene beta-cyclase (lcyB) K06443 [EC:5.5.1.19] mdm-miR159c MD05G1085600 Abscisate beta-glucosyltransferase (AOG) K14595 [EC:2.4.1.263] Table 2.
Pathways detail of miRNAs and target genes involved in volatile organic component synthesis.
Figures
(7)
Tables
(2)