-
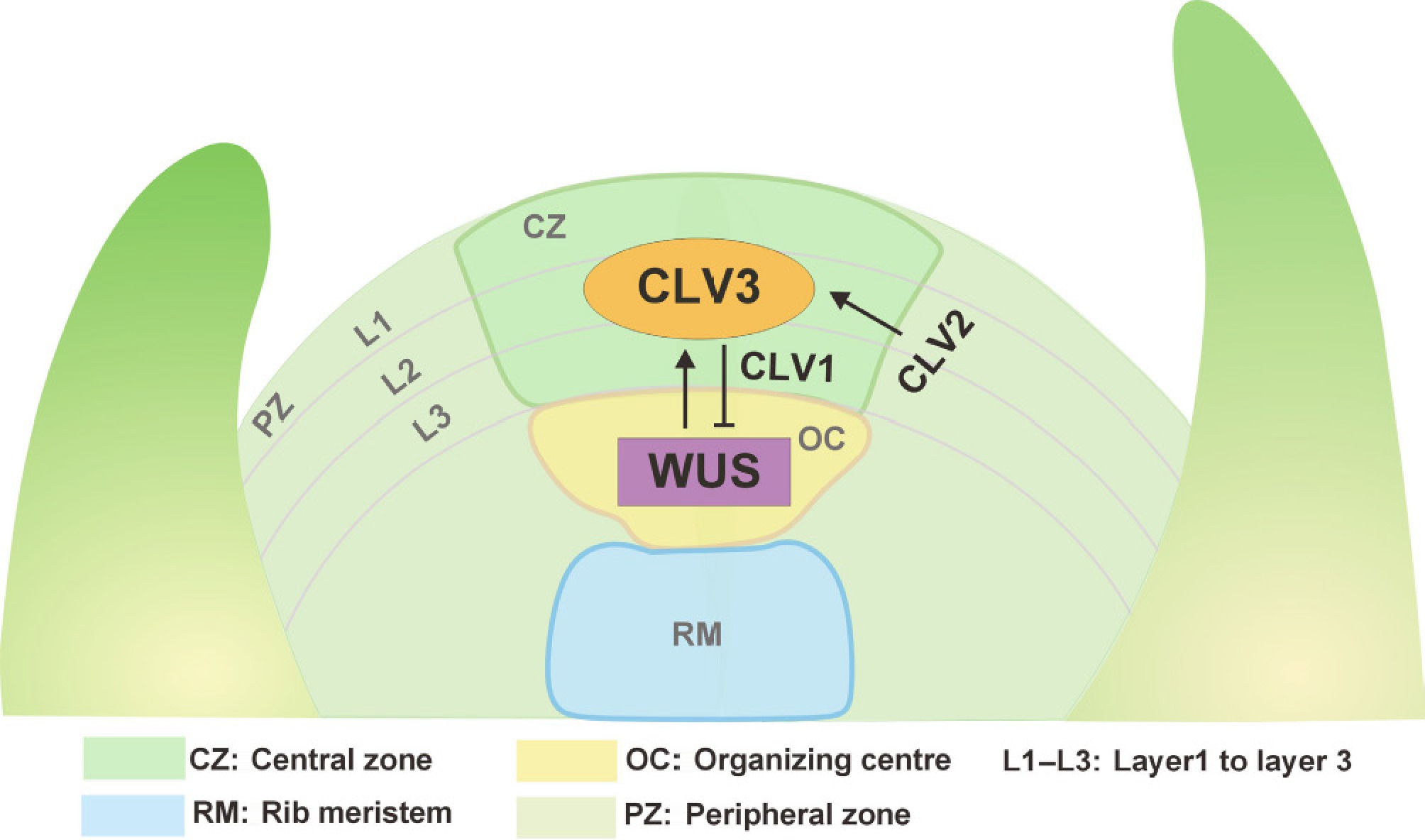
Figure 1.
The CLAVATA3-WUSCHEL negative feedback loop in the shoot meristem. The SAM is composed of three cell layers (L1−L3) and can be divided into distinct functional domains based on its functional and cytological characteristics. These domains include the central zone (CZ), peripheral zone (PZ), organizing center (OC), and rib meristem (RM). The transcription factor WUS is specifically expressed in the OC. Its expression is suppressed by a signaling cascade involving the small peptide CLV3, which binds to the transmembrane receptor kinase CLV1, and the receptor-like protein CLV2. Under normal conditions, the WUS protein moves from the OC to the CZ, where it activates CLV3 expression to inhibit its own activity, thereby ensuring proper growth volume in the plant. Consequently, these components form a feedback loop within the SAM that balances stem cell maintenance and cell differentiation.
-
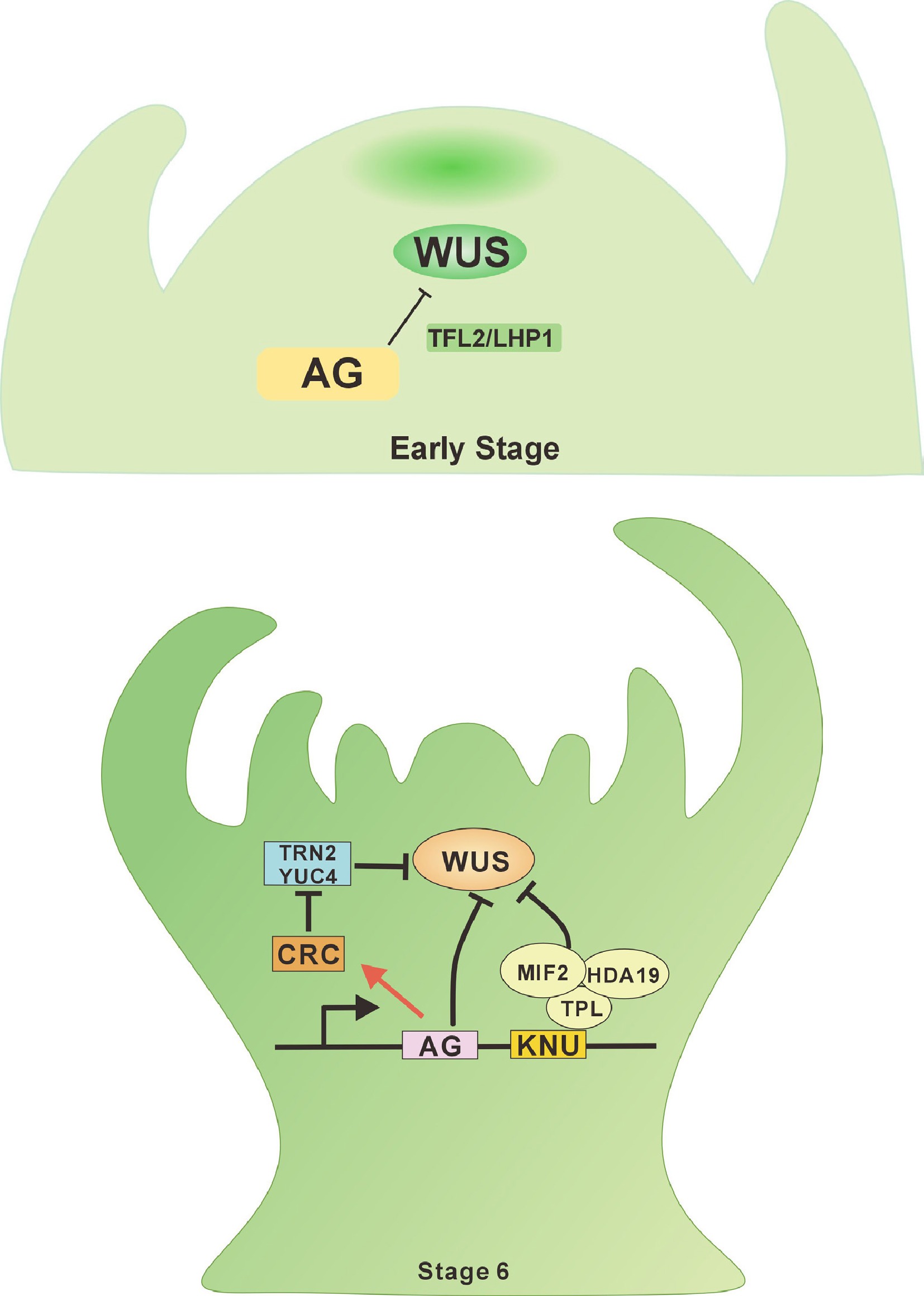
Figure 2.
AGAMOUS-WUSCHEL signaling in the flower meristem. During the early stages of floral development, AG directly represses the WUS gene by recruiting the Polycomb Group (PcG) protein TFL2/LHP1 in the initial phase of floral meristem (FM) termination, thereby terminating floral stem cell fate. At stage 6 of floral development, AG activates the expression of CRC and KNU, which indirectly suppress WUS, ensuring FM determinacy.
-
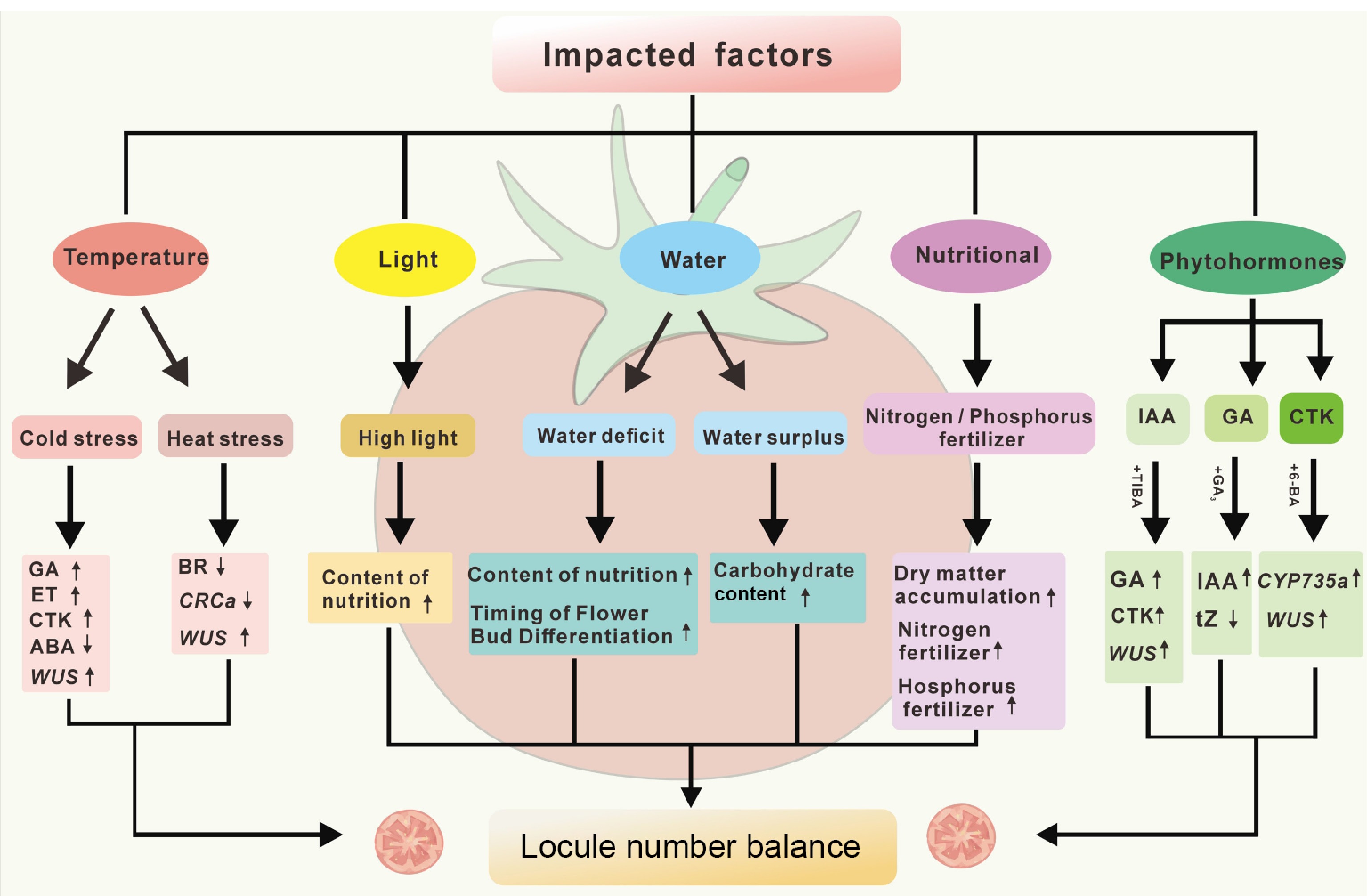
Figure 3.
Critical factors in abnormal fruit development. This diagram summarizes the key factors influencing tomato malformed fruit formation (temperature, light, water, nutrients, and phytohormones), which lead to changes in the levels of GA (gibberellins), CK (cytokinins), ET (ethylene), ABA (abscisic acid), tZ (trans-zeatin), WUS (wuschel), and CRCa (CRABS CLAWa). The upward and downward arrows indicate the increase or decrease of hormone levels, gene expression, or physiological indicators under specific conditions.
-
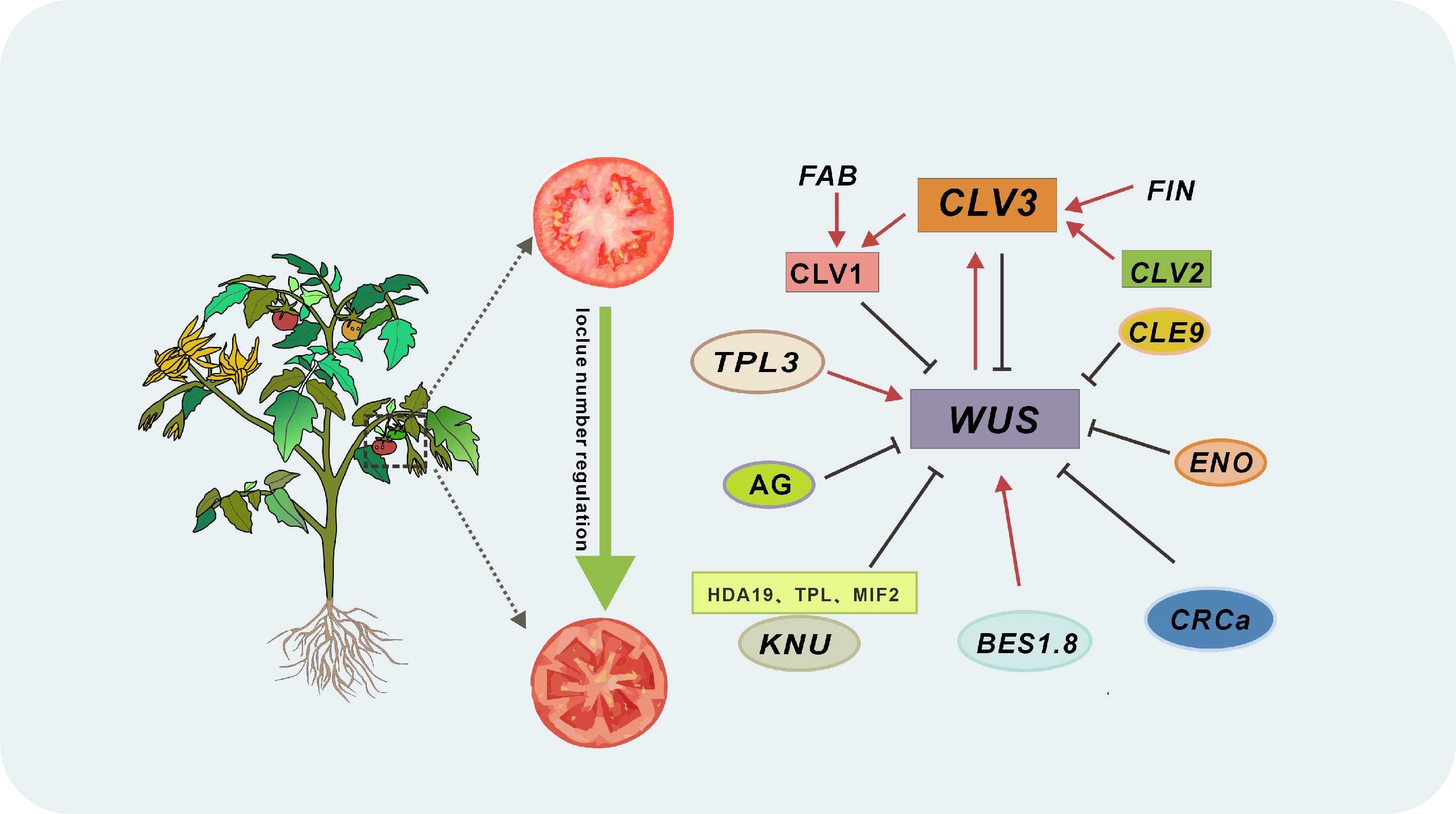
Figure 4.
Integrated networks controlling locule number in tomato. In the plant stem cell regulatory network, CLV1 (encoded by FAB) acts as the receptor kinase for the CLV3 peptide, FIN encodes an arabinosyltransferase that modifies CLV3, collaborating with CLV1 to form a core pathway suppressing WUS activity. When CLV3 is dysfunctional, SlCLE9 activates a partial compensatory mechanism to sustain WUS inhibition. This network integrates multi-tiered transcriptional control: SlENO (an AP2/ERF transcription factor) directly represses SlWUS by binding its promoter; SlBES1.8 sequesters SlWUS via heterodimerization, blocking its interaction with the SlCLV3 promoter and other targets; SlTPL3 partners with WUS to form a transcriptional co-repressor complex, silencing ventricular development-related genes; and AG employs a dual strategy, directly inhibiting WUS in early floral stages, then activating CRCa and KNU at stage 6 to enforce cascade suppression. Arrows denote activation; lines indicate inhibition.
-
Locus/gene (gene number) Chromosomal location Mechanism Mutant phenotype SlCLV3 (Solyc11g071380) Chromosome 11 The mutation of the CArG element downstream of SlWUS results in the loss of repressive function,
leading to the upregulation of WUS expression and affecting the WUS-CLV3 pathway.The number of locules increases by 2 to 4. SlWUS (Solyc02g083950) Chromosome 2 A 294 kb inversion upstream of the SlCLV3 gene disrupts the SlCLV3 promoter, and this mutation affects the WUS-CLV3 pathway. The number of locules increases by 6 to 15. Fab (Solyc04g081590) Chromosome 4 The FAB gene encodes CLV1, a receptor kinase for CLV3. Mutations in FAB can suppress the transcription
of SlCLV3, thereby affecting the WUS-CLV3 pathway.Increase the number of locules. Fin (Solyc11g064850) Chromosome 11 The FIN gene encodes an arabinosyltransferase responsible for the post-translational modification of CLV3. Mutations in FIN can suppress the transcription of SlCLV3, thereby affecting the WUS-CLV3 pathway. Increase the number of locules. SlTPL3 (Solyc01g100050) Chromosome 1 SITPL3 and SIWUS regulate the multicentric phenotype by negatively regulating IAA and positively regulating GA. The shoot apical meristem enlarges, and the number of locules increases. SlENO (Solyc03g117230) Chromosome 3 ENO can interact with the GGC-box cis-regulatory element in the promoter region of SlWUS, directly regulating the expression of SlWUS. The number of flower organs and fruit locules increase. SlIMA (Solyc02g087970) Chromosome 2 SlIMA can assemble with SlKNU, SlTPL1, and HAD19 to form a transcriptional repression complex that suppresses WUS expression. Increase the number of locules. SIBES1.8 (Solyc10g76390) Chromosome 10 SlBES1.8 suppresses the DNA-binding ability of SlWUS by forming a heterodimer through interaction
with it.The shoot apical meristem enlarges, and the number of locules increases. SlKNU (Solyc02g160370) Chromosome 2 SlKNU can assemble with SlMIF2, SlTPL1, and HAD19 to form a transcriptional repression complex that suppresses WUS expression. The number of flower organs and fruit locule increase. SlCLE9 (Solyc06g074060) Chromosome 6 SlCLE9 compensates for the loss of SlCLV3 function by binding to SlWUS, and its mutation exacerbates
the phenotypic defects in SlCLV3 mutants.Increase the number of locules. SlCRCa (Solyc01g010240) Chromosome 1 SlCRCa and SlCRCb bind to chromatin remodeling complex components, thereby suppressing SlWUS expression and promoting floral meristem determinacy. Increase the number of locules. SlCRCb (Solyc05g012050) Chromosome 5 SlCRCa and SlCRCb bind to chromatin remodeling complex components, thereby suppressing SlWUS expression and promoting floral meristem determinacy. Increase the number of locules. Table 1.
Determinants regulating locule number in tomato.
Figures
(4)
Tables
(1)