-
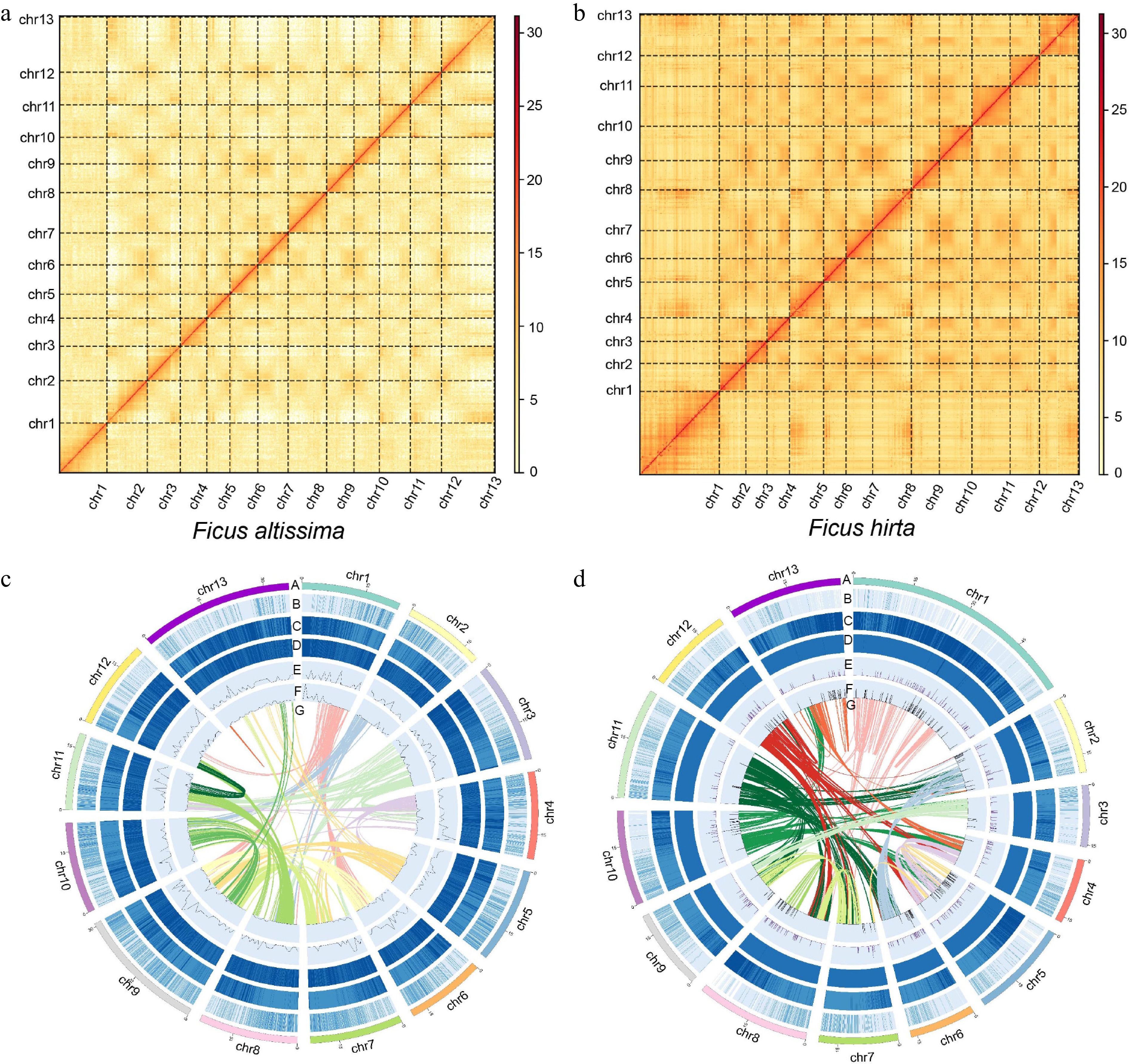
Figure 1.
The Hi-C results and genomic characteristics of Ficus. (a) Hi-C results of F. altissima. (b) Hi-C results of F. hirta. (c) The Circos plot showing the genome details of F. altissima: A, thirteen chromosomes of F. altissima; B, gene density; C, repeat sequence content; D, GC content density; E, density of Copia LTR–RTs; F, density of Gypsy LTR–RTs; G, syntenic blocks (all window sizes = 50 kb). (d) The Circos plot showing the genome details of F. hirta. A, B, C, D, E, F, G: A, thirteen chromosomes of F. hirta; B, gene density; C, repeat sequence content; D, GC content density; E, density of Copia LTR–RTs; F, density of Gypsy LTR–RTs; G, syntenic blocks (all window sizes = 50 kb).
-
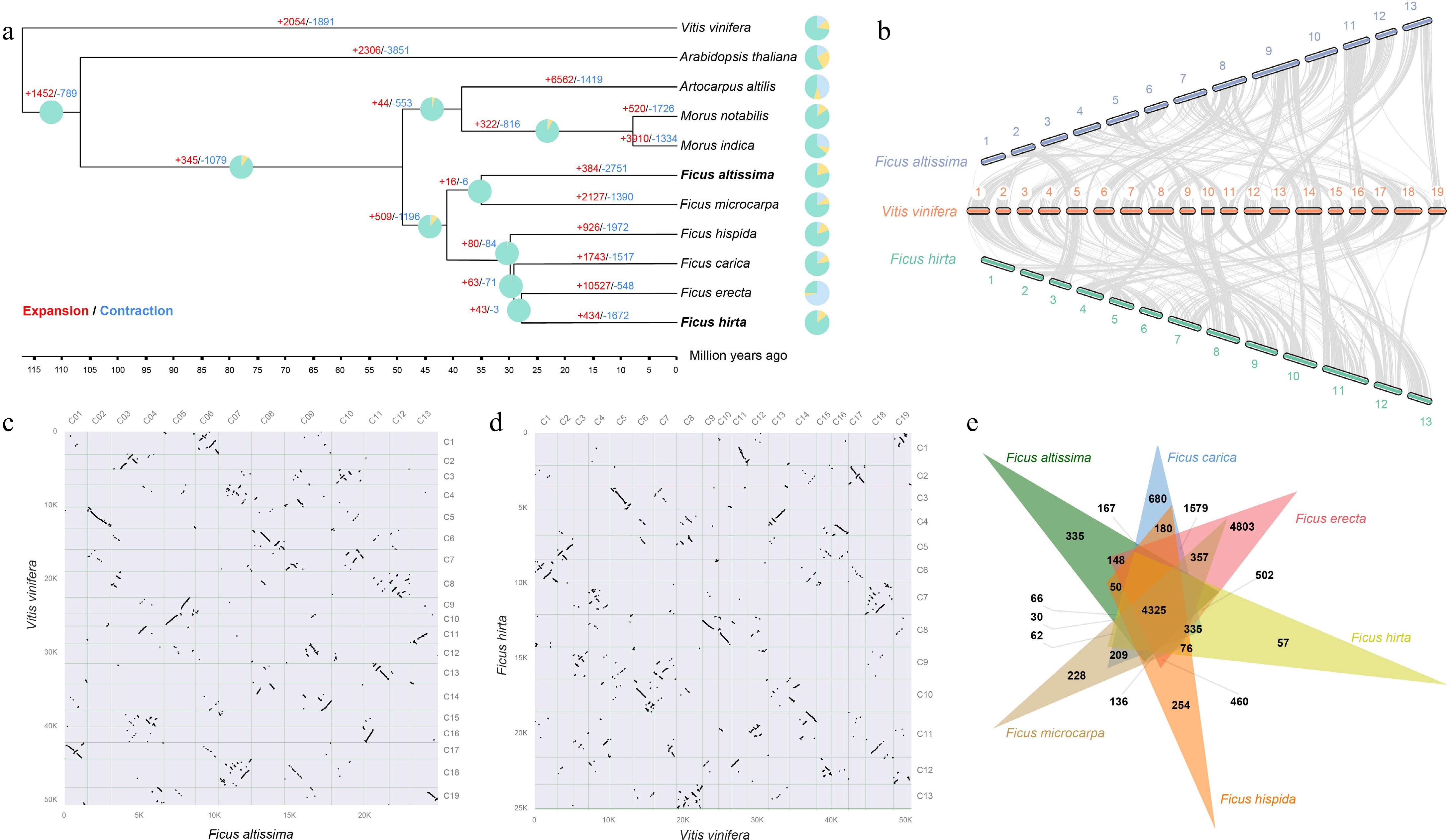
Figure 2.
Evolutionary analysis of the Ficus genus and the genomes of F. altissima and F. hirta. (a) Phylogenetic tree showing F. altissima, F. hirta, and nine other species, along with divergence times and the expansion and contraction of gene families. Green represents gene retention, yellow represents gene contraction, and blue represents gene expansion. (b) Synteny analysis between F. altissima, F. hirta, and Vitis vinifera. (c) Dot plot of F. altissima and V. vinifera. (d) Dot plot of F. hirta and V. vinifera. (e) Venn diagram showing the shared orthologous gene groups among the genomes of the Ficus species.
-
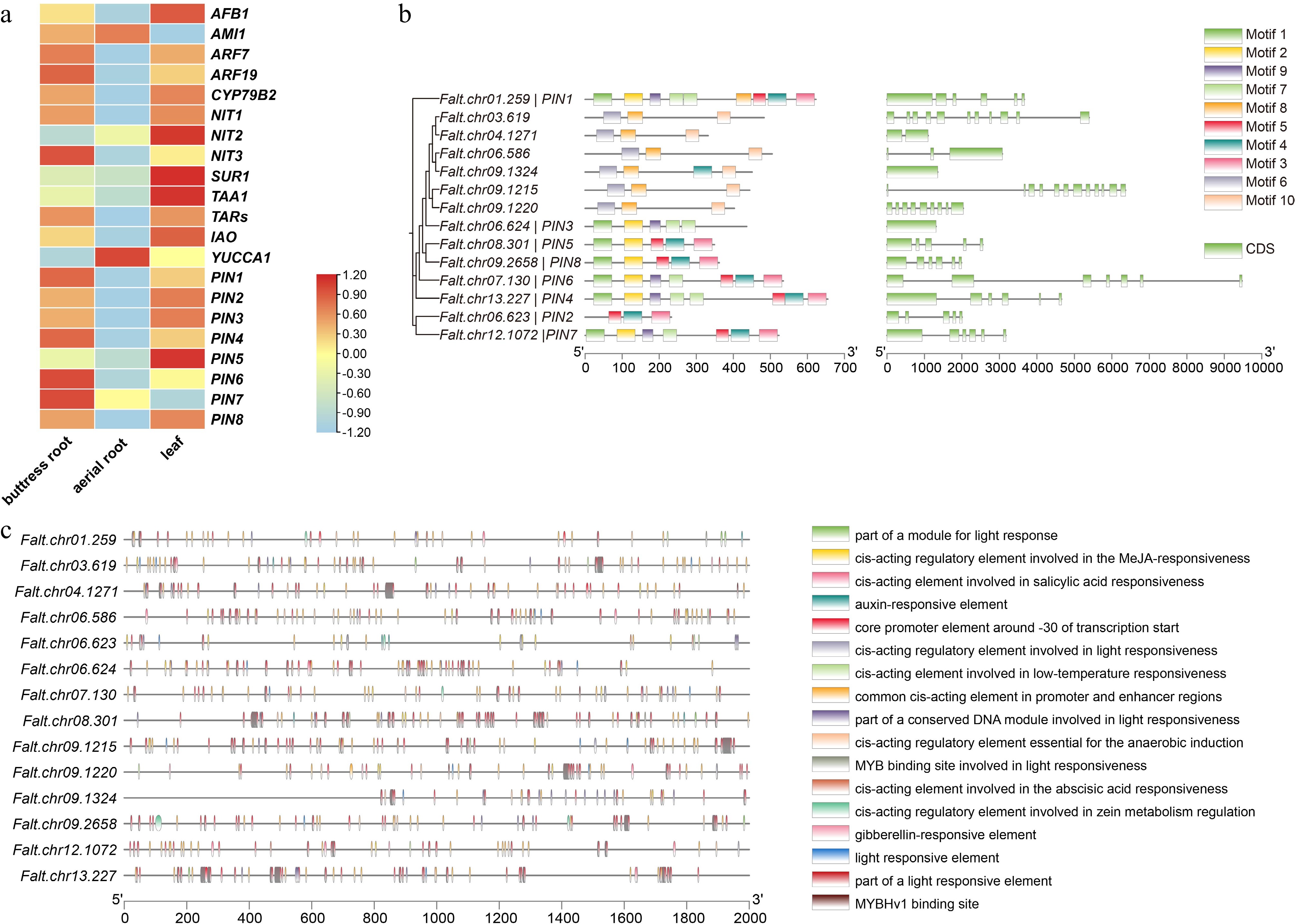
Figure 3.
Characteristics of the PIN gene family in F. altissima. (a) Heatmap of auxin-related genes in F. altissima. (b) Phylogenetic relationships, conserved motifs, and gene structure of the PIN gene family in F. altissima. (c) The role of cis-elements in the PIN gene family of F. altissima.
-

Figure 4.
(a) Total metabolites extracted from the thin root, medium root, and course root. (b) Differentially expressed top 20 metabolites between thin root and course root, showing that psoralen is highly expressed in the course root.
-
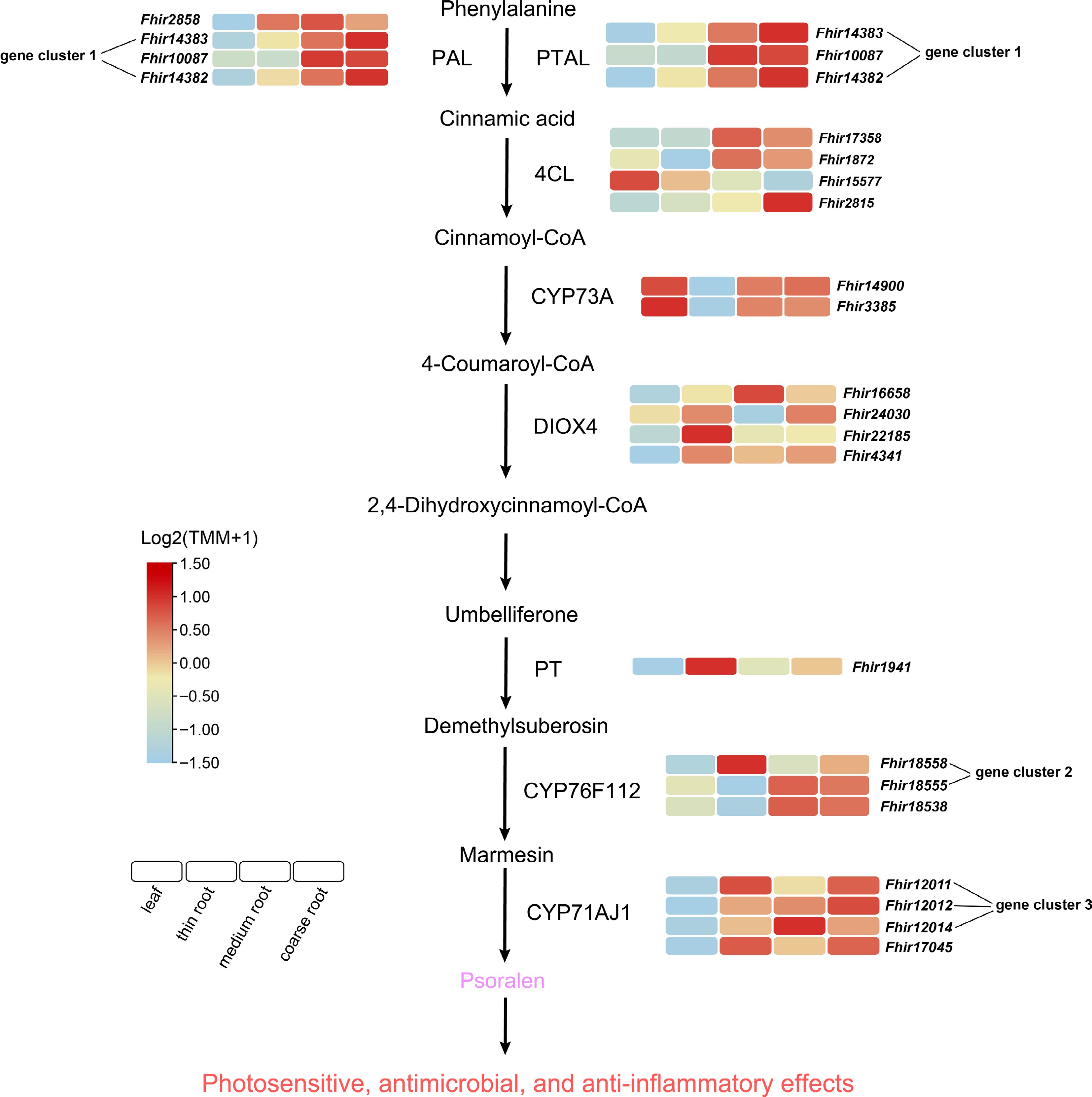
Figure 5.
Biosynthetic pathway of psoralen and gene expression in F. hirta. PT: phenylalanine ammonia-lyase, PTAL: phenylalanine/tyrosine ammonia-lyase, 4CL: 4-coumarate--CoA ligase, CYP73A: trans-cinnamate 4-monooxygenase, DIOX4: trans-4-coumaroyl-CoA 2-hydroxylase, PT: umbelliferone 6-dimethylallyltransferase, CYP76F112: marmesin synthase, CYP71AJ1: psoralen synthase.
-
Genomic feature F. altissima F. hirta Genome size (Mb) 355.9 323.7 Number of chromosomes 13 13 Scaffold N50 length (Mb) 22.8 24.8 GC content (%) 35.21 34.73 Genomic heterozygous (%) 1.33 1.02 Repeat sequence content (%) 54.11 52.14 The number of protein-coding genes 24,938 25,170 Genome BUSCOs (%) 98.2 98.2 Merqury completeness 86.96 76.88 Table 1.
Basic genomic information of F. altissima and F. hirta.
Figures
(5)
Tables
(1)