-
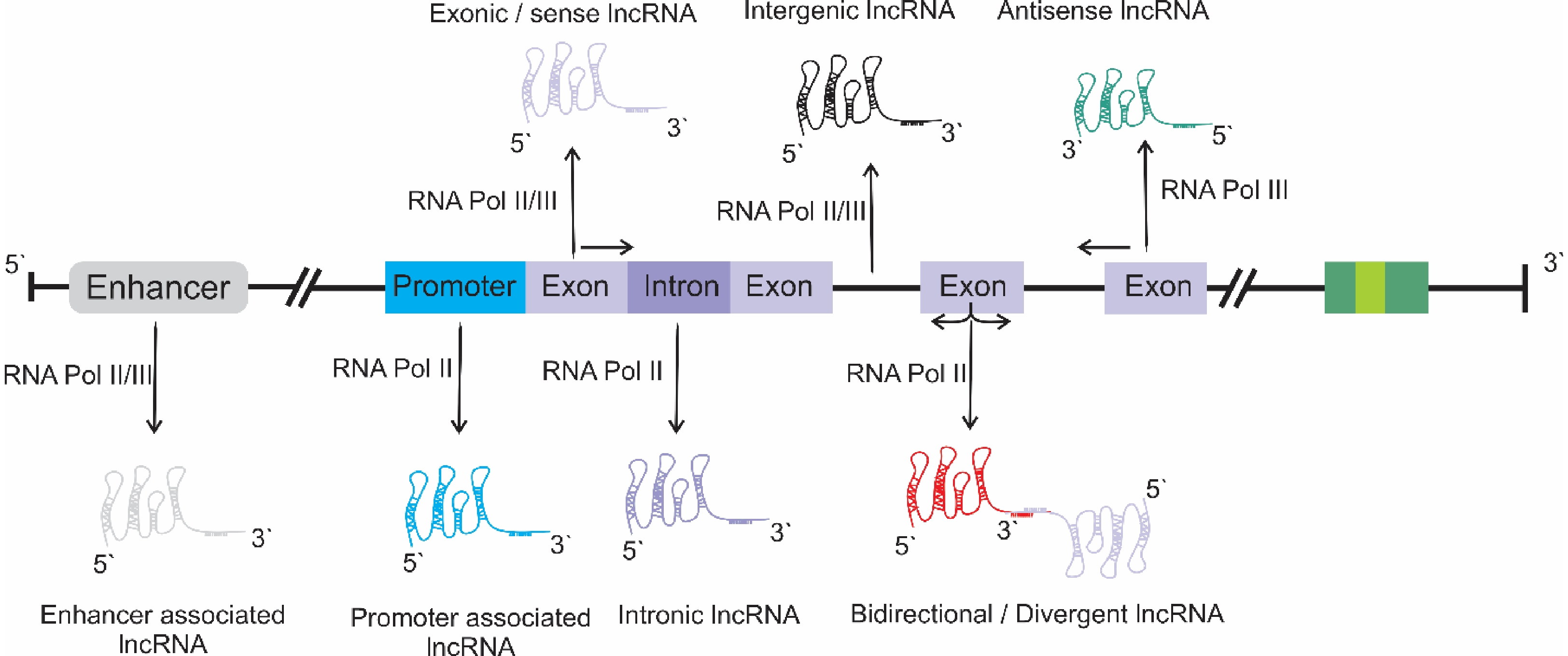
Figure 1.
Different classes of lncRNAs, based on their origin in the genome context.
-
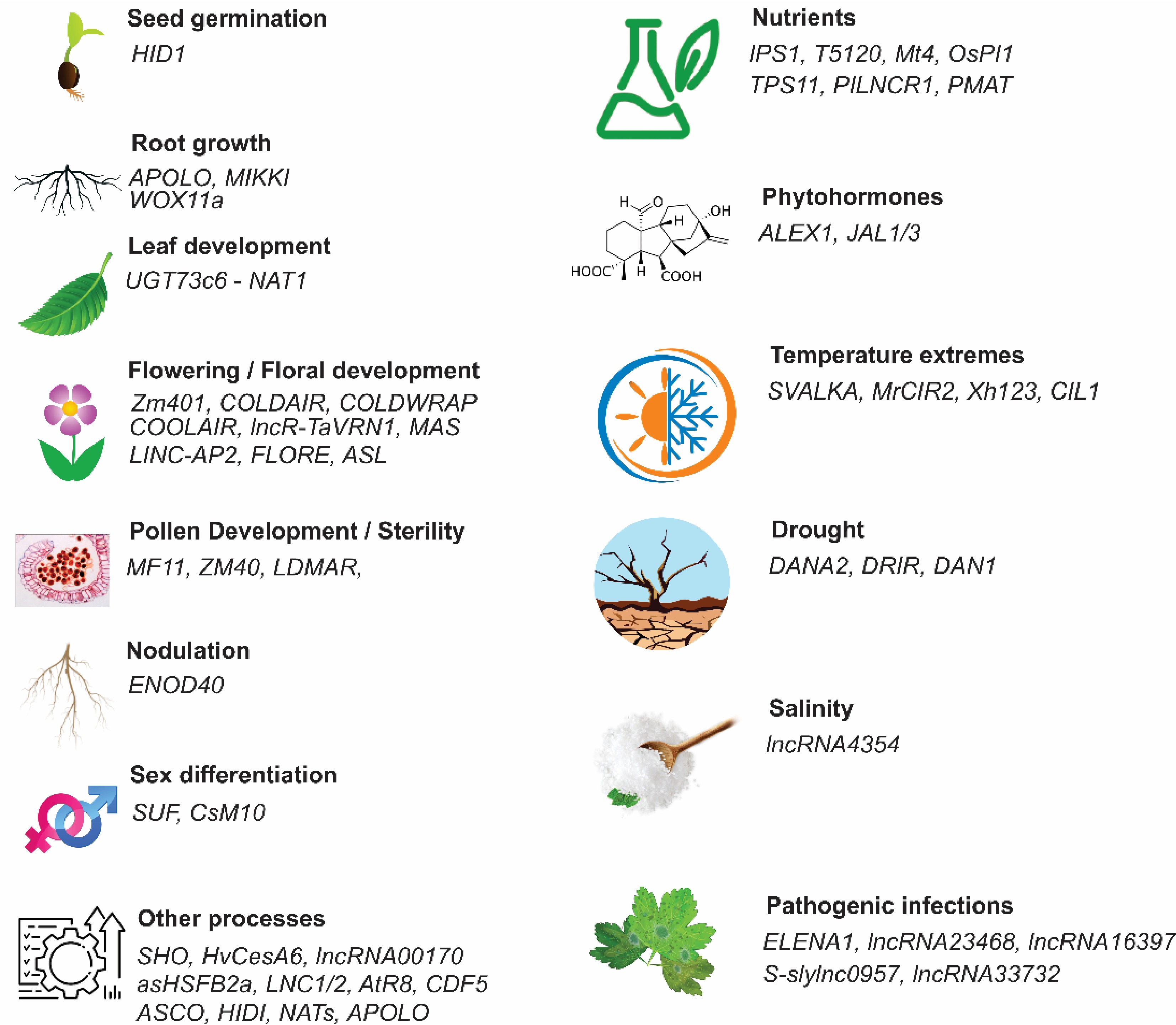
Figure 2.
Functional regulation of long non-coding RNAs (lncRNAs) in plant growth, development, and stress responses. HID1 plays an important role in seed germination, APOLO, MIKKI, and WOX11a alter root development and root system architecture, leaf morphogenesisis controlled by UGT73c6 (a NAT lncRNA) is regulated cold and spring flowering, flowering time by Zm401, COLDAIR, COLDWRAP, COOLAIR, lncR-TaVRN1, MASS, LINC-AP2, FLORE, and ASL, pollen development is control by MF11, ZM40, and LDMAR. ENOD40 promotes root nodulation. Sex differentiation is determined by SUF and CsM10. Plant ability to enhance nutrient accumulation is in addition to a protein-coding genes is also contributed by lncRNAs like IPS1, T5120, Mt4, OsPI1, TPS11, PILNCR1, and PMAT. SVALKA, MrCIR2, Xh123, and CIL1 regulate tolerance to high temperatures, while DANA2, DRIR, and DAN1 regulate drought tolerance, and lncRN4354 regulates salinity stress in plants. ALEX1 and JAL1/3 regulate phytohorme-mediated responses in plants. However, there are several lncRNAs like ELENA1, lncRNA23468, lncRNA16397, S-sylnc0957, and lncR33732 that regulate pathogenic infections and enhance disease resistance[1].
-
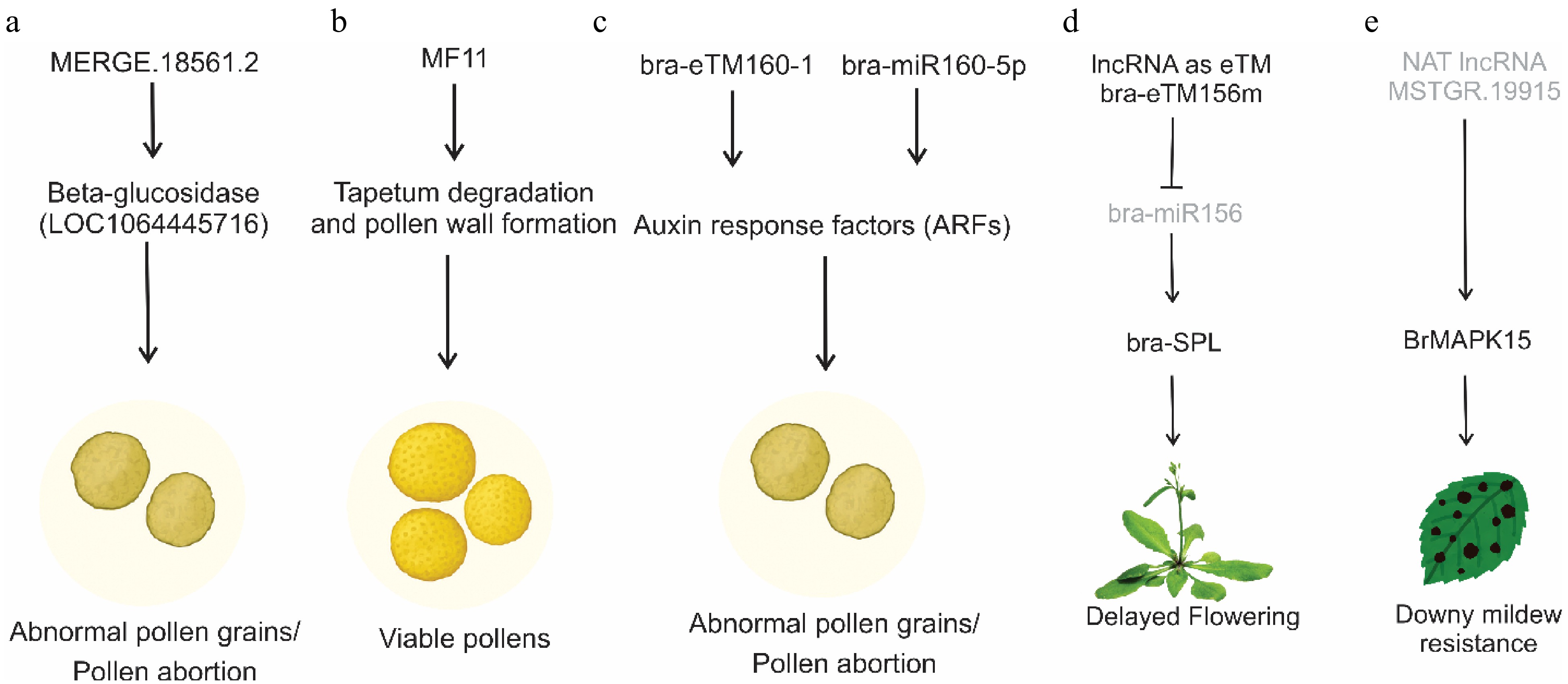
Figure 3.
Mechanisms of action of lncRNAs in different developmental and stress responses. (a) a lncRNA (MERGE.18561.2) impacts pollen viability by targeting beta-glucosidase genes. (b) MF11 suppresses pollen germination efficiency, delays pollen tube germination, delays tapetum degradation, and aborts the development of pollen grains. (c) lncRNAs acting as endogenous targets of bra-miR160, bra-eTM160-1, and bra-eTM160-2, suppress the activity of miR160 activating ARF genes, which subsequently influence pollen development. (d) lncRNA as eTM of bra-miR156 in Chinese cabbage, which ultimately delays flowering by activating SPL genes. (e) An antisense lncRNA activates the MAPK15 gene to enhance resistance against downy mildew.
-
Brassica plant Number of lncRNAs Developmental stage Stimuli / environmental condition Ref. Abiotic Biotic Brassica rapa 1,031 NAT-lncRNAs Heat [12] 12,051 lncRNAs Pollen development and fertilization [13] 18,253 lncRNA Heat [14] 254 DE-lncRNAs Vernalization [15] 407 lncRNAs Anthers development [16] 278 lncRNAs Heat [17] 4,594 lncRNAs Heat [18] 6,392 lncRNAs Pollen development [19] 385 DE-lncRNAs Male sterility and fertility [20] Brassica napus 3,181 lncRNA Sclerotinia sclerotiorum [21] 11,073 lncRNAs Cold [22] 8,094 lncRNAs Seed oil accumulation 5,546/6,997 in Q2 and 7,824/10,251 Qinyou8 down/upregulated Drought and re-watering [23] 2,546 lncRNAs Cadmium [24] Brassica juncea 7,613 lncRNAs with 1,614 DE-lncRNAs Drought and heat stress [25] 3,602 DE-lncRNAs Salinity [26] 1,539 lncRNAs Seed development [27] Brassica oleraceae 148 DE-lncRNAs Cuticular Wax Biosynthesis [28] Table 1.
Genome-scale identification of lncRNAs in Brassica crops in specific tissues, developmental stages, and environmental conditions.
Figures
(3)
Tables
(1)