-
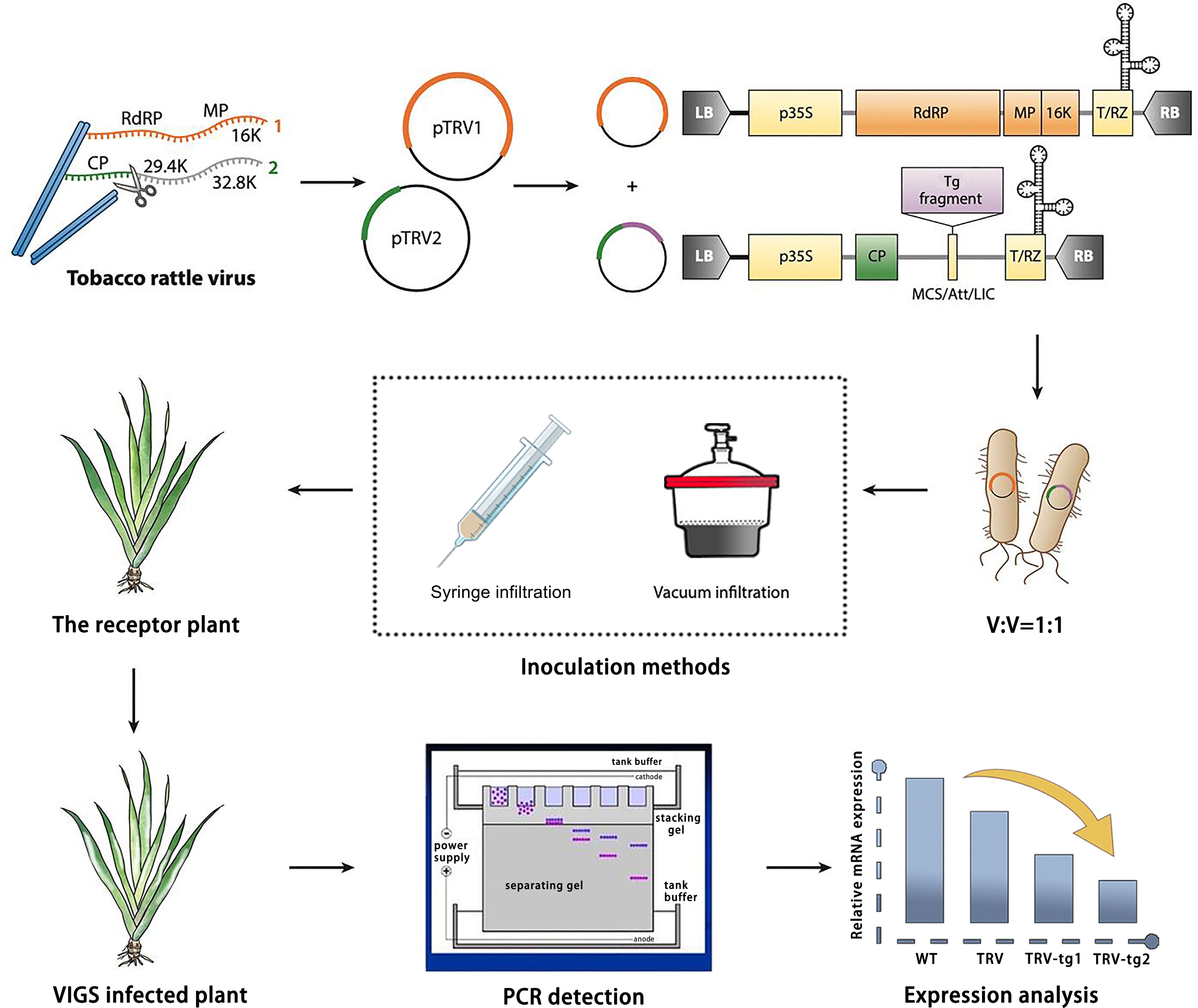
Figure 1.
Design and application of TRV-based virus-induced gene silencing (VIGS) in Iris japonica. TRV has a bipartite genome (genome 1 in orange; genome 2 in green). Genome 1 is incorporated into a vector backbone without modification, while only the coat protein (CP) for genome 2 is essential for VIGS. TRV vectors, like the binary pTRV1/pTRV2 system, are typically introduced into plants via Agrobacterium tumefaciens inoculation, with T-DNA borders (LB and RB) added. Strong promoters (e.g., p35S or 2×p35S) drive viral genome expression (yellow boxes show required elements for transcription). The vector may also include terminators (e.g., NOS), a self-cleaving ribozyme (T/RZ), and a multiple cloning site (MCS) or recombination system for reliable insertion cloning. For TRV-based VIGS, a target (e.g., IjPDS) gene fragment is inserted into the vector, which is then used to infect the receptor material via A. tumefaciens. Afterwards, the phenotype is observed, and gene function is validated by the phenotype observation (photobleached leaves), PCR detection, and gene expression analysis.
-
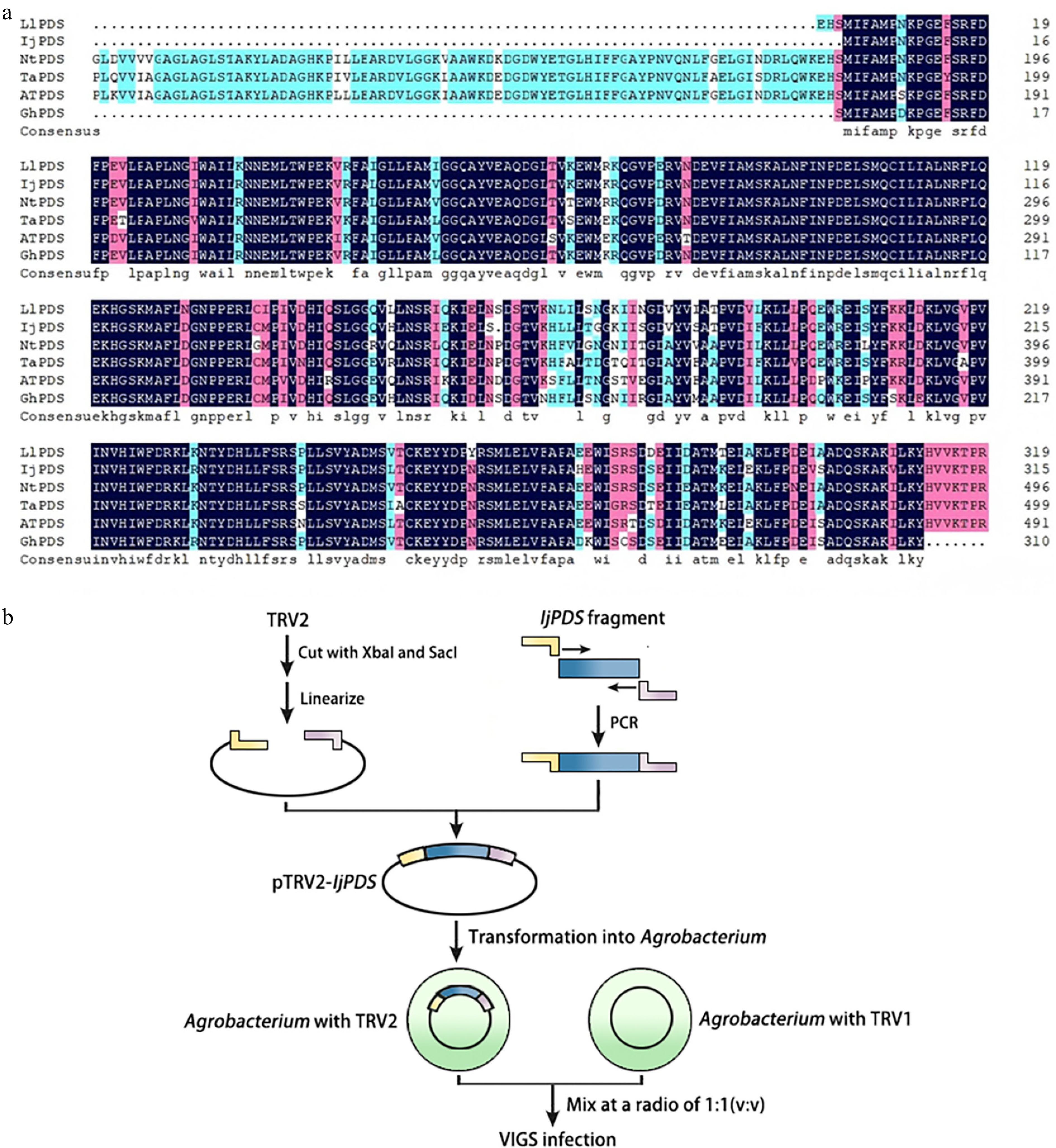
Figure 2.
Identification of IjPDS and construction of VIGS vector (pTRV2-IjPDS). (a) Multiple sequence alignment of the PDS gene from Iris japonica, a model plant, and related monocot species to identify conserved sequences. LlPDS: Lilium longiflorum, BAV93014.1; IjPDS: Iris japonica, T02.PB9671; NtPDS: Narcissus tazetta, AFH53816.1; TaPDS: Triticum aestivum, ACL36586.1; AtPDS: Arabidopsis thaliana, AT4G14210; GhPDS: Gladiolus hybrid, AGI17588.1. The red rectangles indicate the target fragment used for silencing IjPDS. (b) Procedure for pTRV2-IjPDS construction and agro-infiltration. The yellow and pink lines represent adaptors used in ligation-independent cloning, while the blue lines represent the target IjPDS fragment.
-

Figure 3.
Agrobacterium tumefaciens inoculation of Iris japonica receptor plants in the VIGS System. (a) Rhizomes and clipped leaves of receptor plants pricked with toothpicks. (b) Receptor plants placed in a beaker for inoculation with the prepared A. inoculation solution. (c) Receptor plants subjected to vacuum infiltration to enhance infection efficiency. (d) Photobleached phenotype observed in the newly developed leaves of silenced plant after two weeks.
-
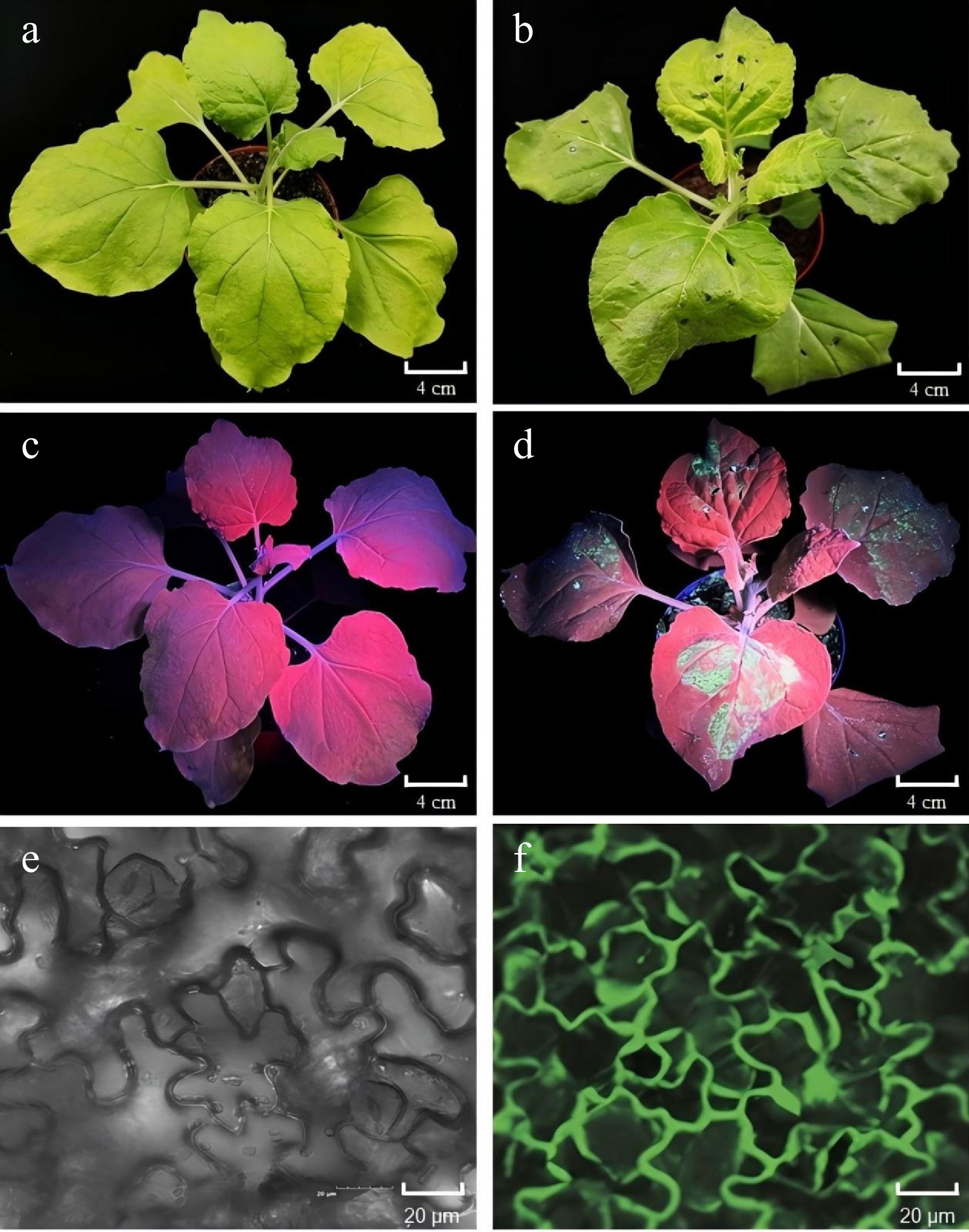
Figure 4.
Validation of the pTRV2-GFP vector in tobacco (Nicotiana tabacum) leaves. (a), (b) Photographs of tobacco leaves taken 6 d post-infiltration (dpi) without or with pTRV2-GFP, captured under open-field lighting conditions. (c), (d) GFP expression observed under ultraviolet (UV) light in tobacco leaves at 6 dpi, without and with pTRV2-GFP. GFP fluorescence was detected in regions corresponding to GFP expression in the treated areas. (e), (f) GFP fluorescence detected using a laser scanning confocal microscope (Olympus FV3000, Kyoto, Japan) in tobacco leaves at 6 dpi without or with pTRV2-GFP. Fluorescent regions indicate GFP expression in the treated leaves.
-
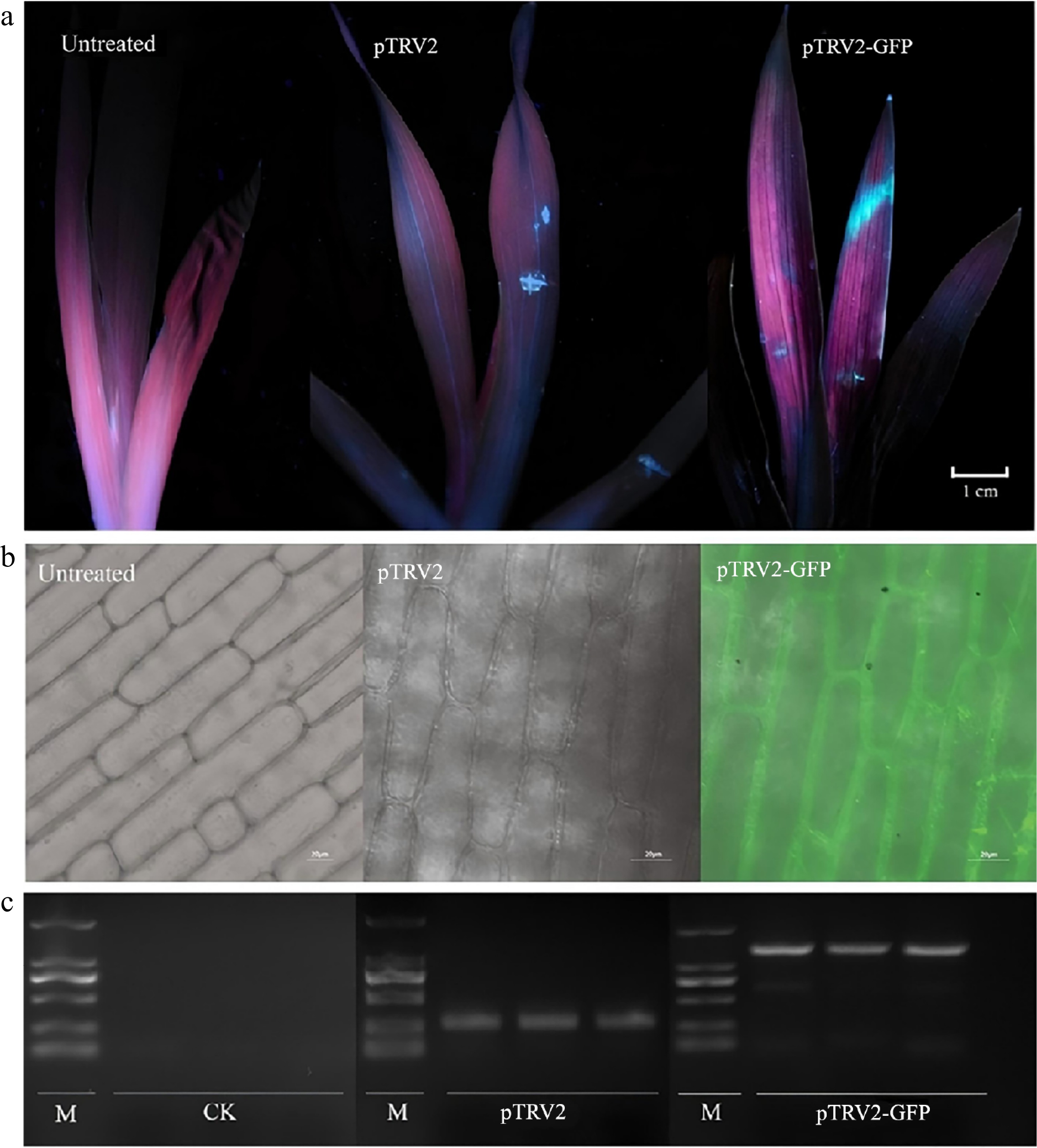
Figure 5.
Application of the pTRV2-GFP vector in Iris japonica. (a) GFP expression observed under ultraviolet (UV) light in I. japonica leaves 7 d post-infiltration (dpi), comparing untreated, pTRV2-treated, and pTRV2-GFP-treated leaves. UV fluorescence was detected in the regions on the leaves treated with pTRV2 and pTRV2-GFP, confirming GFP expression. (b) GFP expression observed under a laser scanning confocal microscope (Olympus FV3000, Kyoto, Japan) in I. japonica leaves at 7 dpi, comparing untreated, pTRV2-treated and pTRV2-GFP-treated leaves, confirming successful expression. GFP fluorescence was detected in the regions of the pTRV2-GFP-treated leaves, confirming successful expression. (c) PCR validation of GFP expression in I. japonica leaves 7 dpi, comparing untreated, pTRV2-treated, and pTRV2-GFP-treated leaves. A GFP-specific band was observed in the pTRV2-GFP-treated leaves, indicating the successful inoculation of pTRV2 and pTRV2-GFP vectors in I. japonica.
-
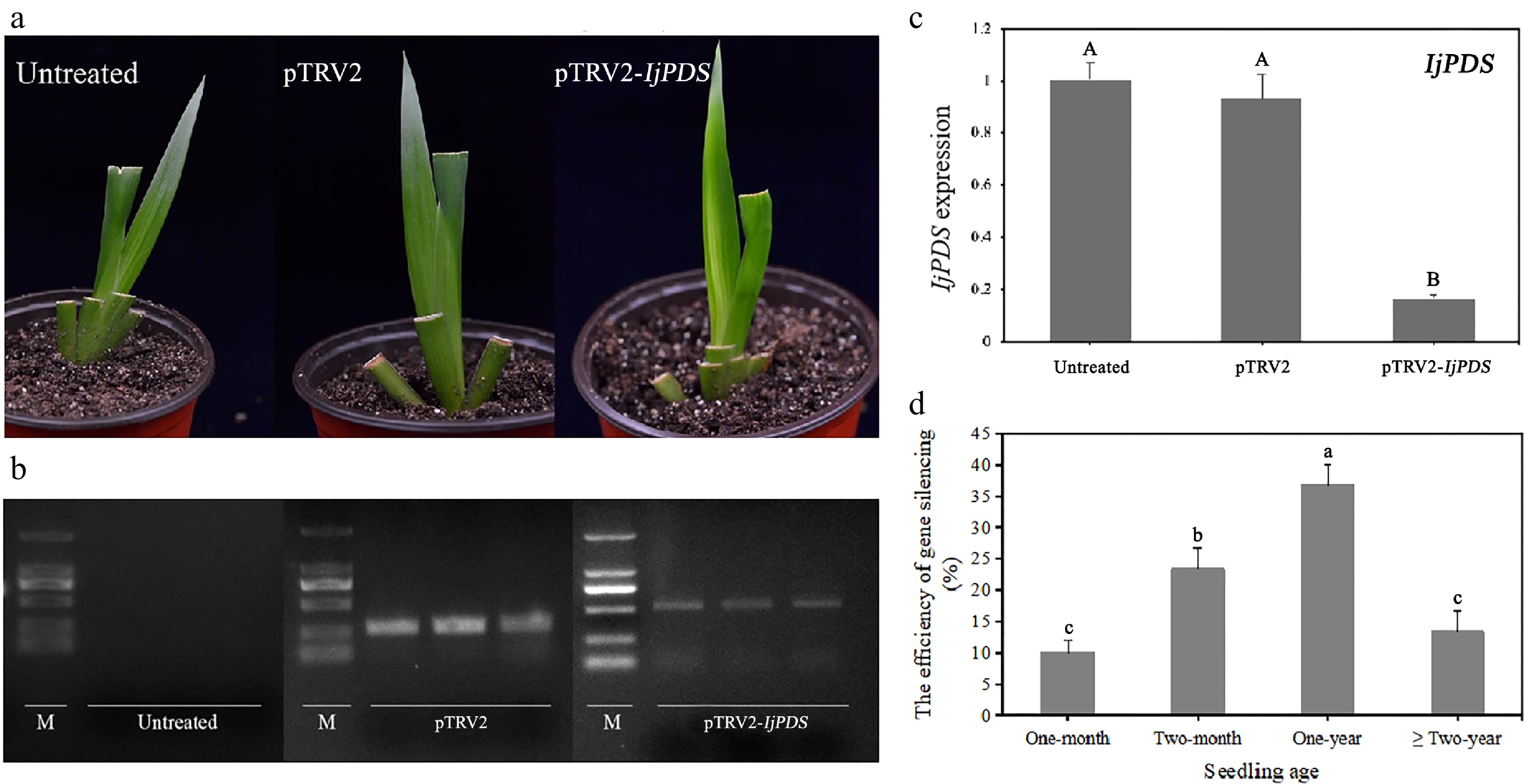
Figure 6.
Phenotype and molecular validation of IjPDS silencing in Iris japonica. (a) Phenotypic observations of untreated, pTRV2-treated, and pTRV2-IjPDS-treated I. japonica leaves at 28 d post-infiltration (dpi). A characteristic photobleached phenotype was observed in the newly developed leaves of pTRV2-IjPDS-treated plants at 28 dpi, indicating successful silencing of IjPDS. (b) RT-PCR analysis of PDS expression in I. japonica leaves at 28 dpi from untreated, pTRV2-treated, and pTRV2-IjPDS-treated plants. A larger specific band was observed in the pTRV2-IjPDS-treated leaves compared to the pTRV2-treated leaves, confirming the infection of the pTRV2-IjPDS vector. In contrast, there is no TRV band detected in untreated plants. (c) Quantitative real-time PCR (qPCR) analysis of PDS expression in I. japonica leaves at 28 dpi from untreated, pTRV2-treated, and pTRV2-IjPDS-treated plants. Reduced PDS expression was detected in the pTRV2-IjPDS-treated plants, confirming successful silencing of IjPDS in I. japonica leaves. (d) Comparison of infection efficiency at different seedling ages, demonstrating the optimal age for successful silencing in I. japonica. Each age group consists of three sets of uniformly growing plants, with ten plants per set. The data represent the mean values ± standard error. Different letters in (c) and (d) indicate significant differences at p < 0.05, following statistical analysis by ANOVA.
-
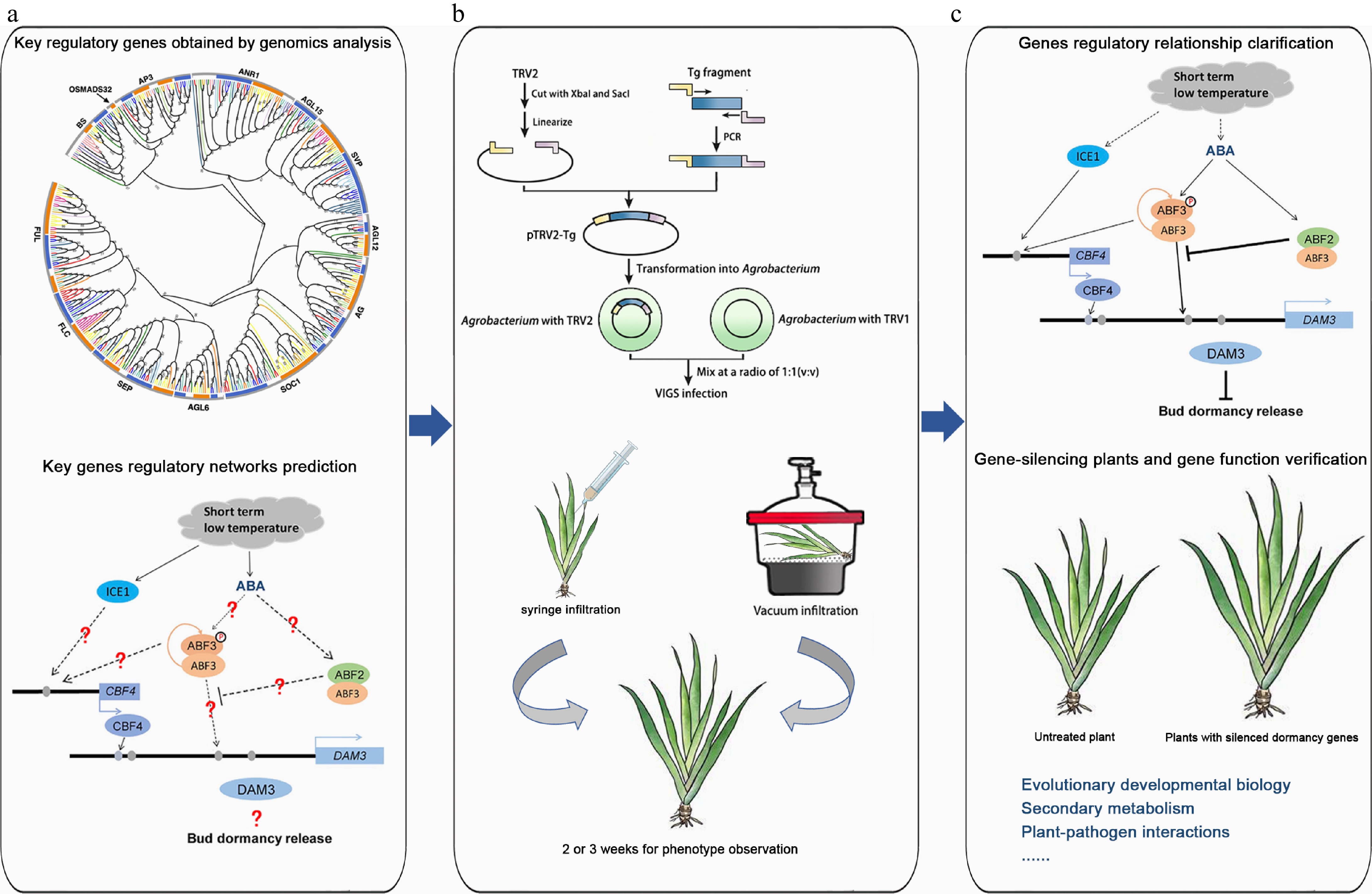
Figure 7.
The application of the TRV-VIGS system in plant research using Iris japonica as an example. (a) Genomics analysis. Key regulatory genes were identified through genomics analysis, and the regulatory networks of these genes were predicted. The graphs of phylogenetic analysis and regulatory network were modified from Li et al.[5] and Yang et al.[57], respectively. (b) Homologous gene verification of homologous gene function by VIGS. A target gene fragment was constructed into the TRV vector, and various Agrobacteria-mediated methods, such as syringe infiltration and vacuum infiltration, were used to infect Iris leaves to verify homologous gene function. (c) Gene function research. The regulatory relationships between genes were explored, gene-silenced plants were obtained, and gene functions were validated. Additionally, VIGS technology is not limited to gene function verification but can also be applied to research in evolutionary and developmental biology, secondary metabolism, immune interactions, and symbiotic relationships.
-
Primer name Sequence (5'–3') IjPDS-F 5'-GCTCTAGAATCACTGGGTGGTCAGGTCC-3' IjPDS-R 5'-CGAGCTCTTCTCAGCTTCCTGTCAAACCA-3' M13-F 5'-GTAAAACGACGGCCAGT-3' M13-R 5'-CAGGAAACAGCTATGAC-3' pTRV2-F 5'-TGGGAGATGATACGCTGTT-3' pTRV2-R 5'-CCTAAAACTTCAGACACG-3' pTRV1-F 5'-TTACAGGTTATTTGGGCTAG-3' pTRV1-R 5'-CCGGGTTCAATTCCTTATC-3' Table 1.
Primers for gene clone and quantitative real-time PCR analysis.
-
Efficiency Vector Group pTRV2-GFP pTRV2-IjPDS pTRV2-GFP-IjPDS 1 60% 30% 30% 2 50% 40% 30% 3 50% 40% 40% Average 53.33% 36.67% 33.33% SD 5.77% 5.77% 5.77% Significant differences indicated by different letters a b b Table 2.
Efficiency assessment of TRV-VIGS system using pTRV2-GFP and pTRV2-IjPDS visual vectors.
Figures
(7)
Tables
(2)