-
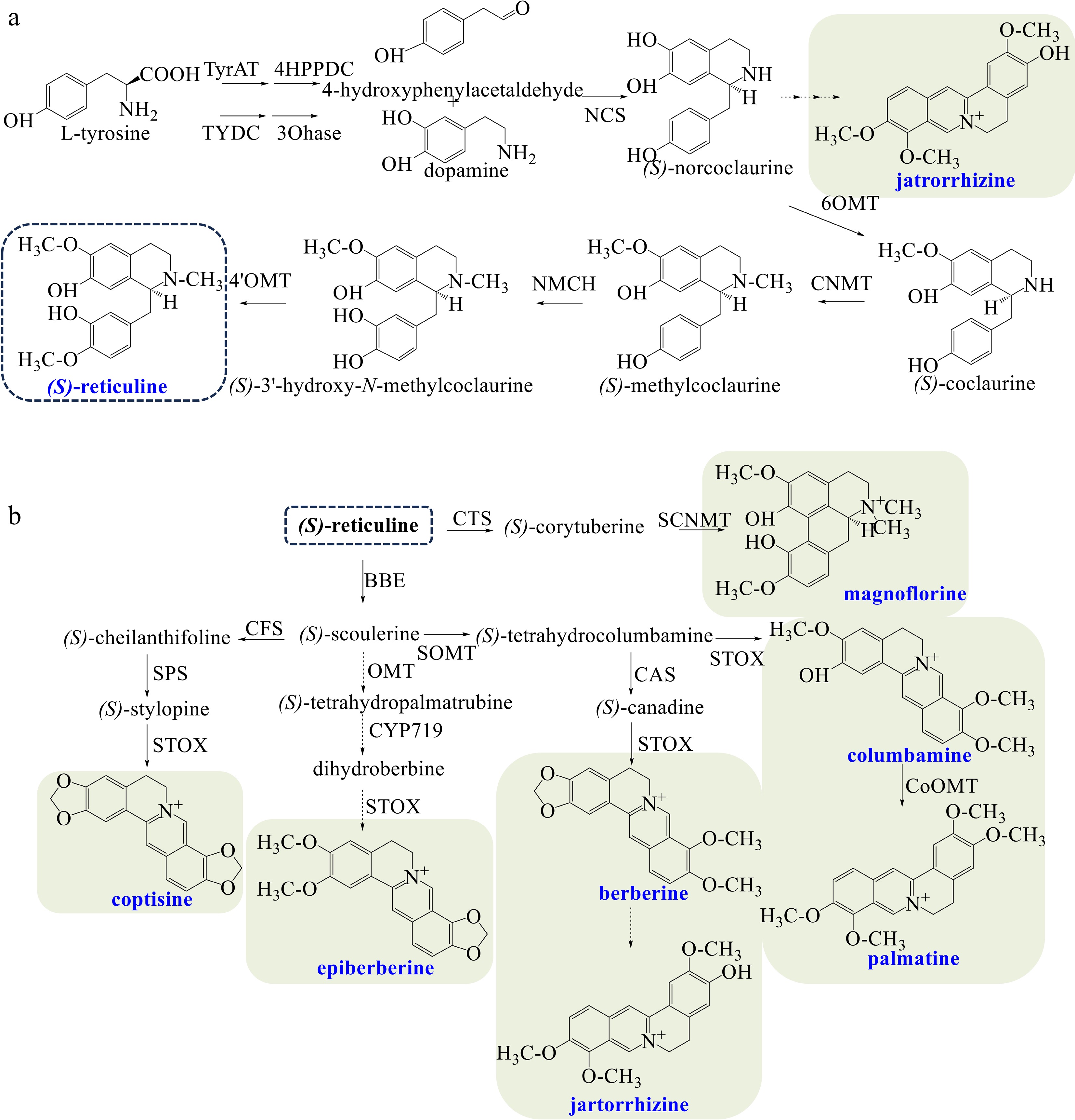
Figure 1.
(a) Biosynthetic pathways of the central intermediate (S)-reticuline. (b) Biosynthetic pathways of diverse BIAs in Coptis species. The broken arrows represent a hypothetical pathway that has not yet been substantiated. Abbreviations: TyrAT, L-tyrosine aminotransferase; 4HPPDC, 4-hydroxyphenylpuruvate decarboxylase tyrosine/tyramine; TYDC, 3-hydroxylase tyrosine decarboxylase; 3OHase, tyrosine/tyramine 3-hydroxylase; NCS, (S)-norcoclaurine synthase; 6OMT, 6-O-methyltransferase; CNMT, (S)-coclaurine N-methyltransferase; NMCH, N-methylcoclaurine 3'-hydroxylase; 4'OMT, 3'-hydroxy-N-methylcoclaurine 4'-O-methyltransferase; CTS, corytuberine synthase; SCNMT, (S)-corytuberine-N-methyltransferase; BBE, berberine bridge enzyme; SOMT, soulerine 9-O-methyltransferase; CAS, canadine synthase; STOX, tetrahydroprotoberberine oxidase; CoOMT, O-methyltransferase; CFS, (S)-cheilanthifoline synthase; SPS, (S)-stylopine synthase.
-
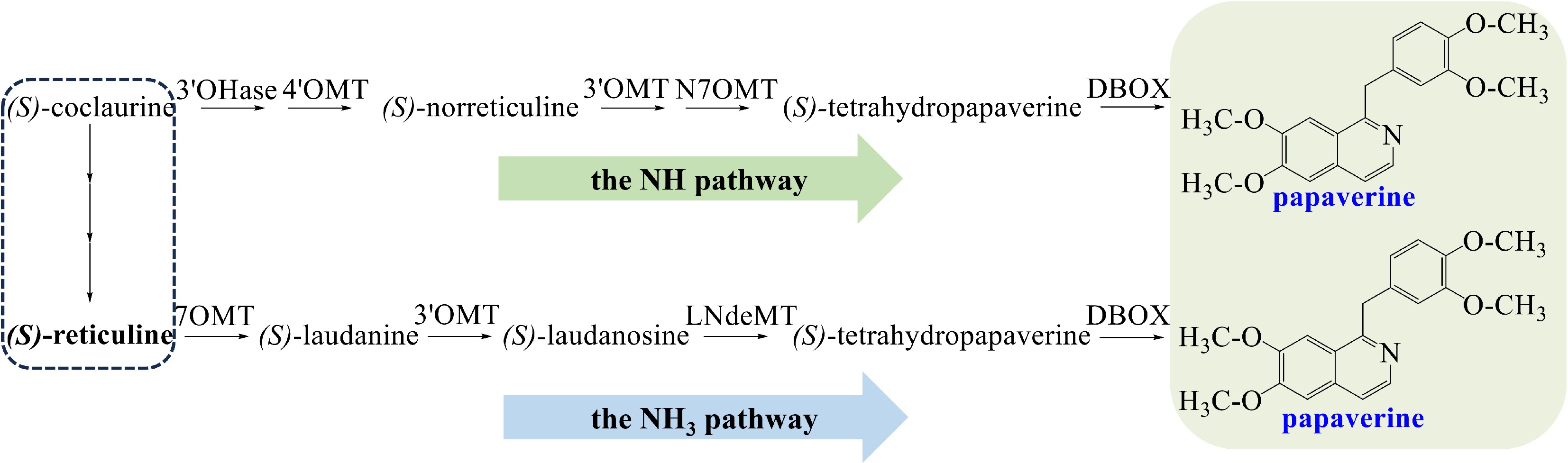
Figure 2.
The NH and NH3 biosynthetic pathway of papaverine. Abbreviations: 3'OHase, 3'-hydroxylase; 4'OMT, 3'-hydroxy-N-methylcoclaurine 4'-O-methyltransferase; 3'OMT, 3'-O-methyltransferase; N7OMT, norreticuline 7-O-methyltransferase; DBOX, dihydrobenzophenanthridine oxidase; 7OMT, 7-O-methyltransferase; LndeMT, laudanosine N-demethylase.
-
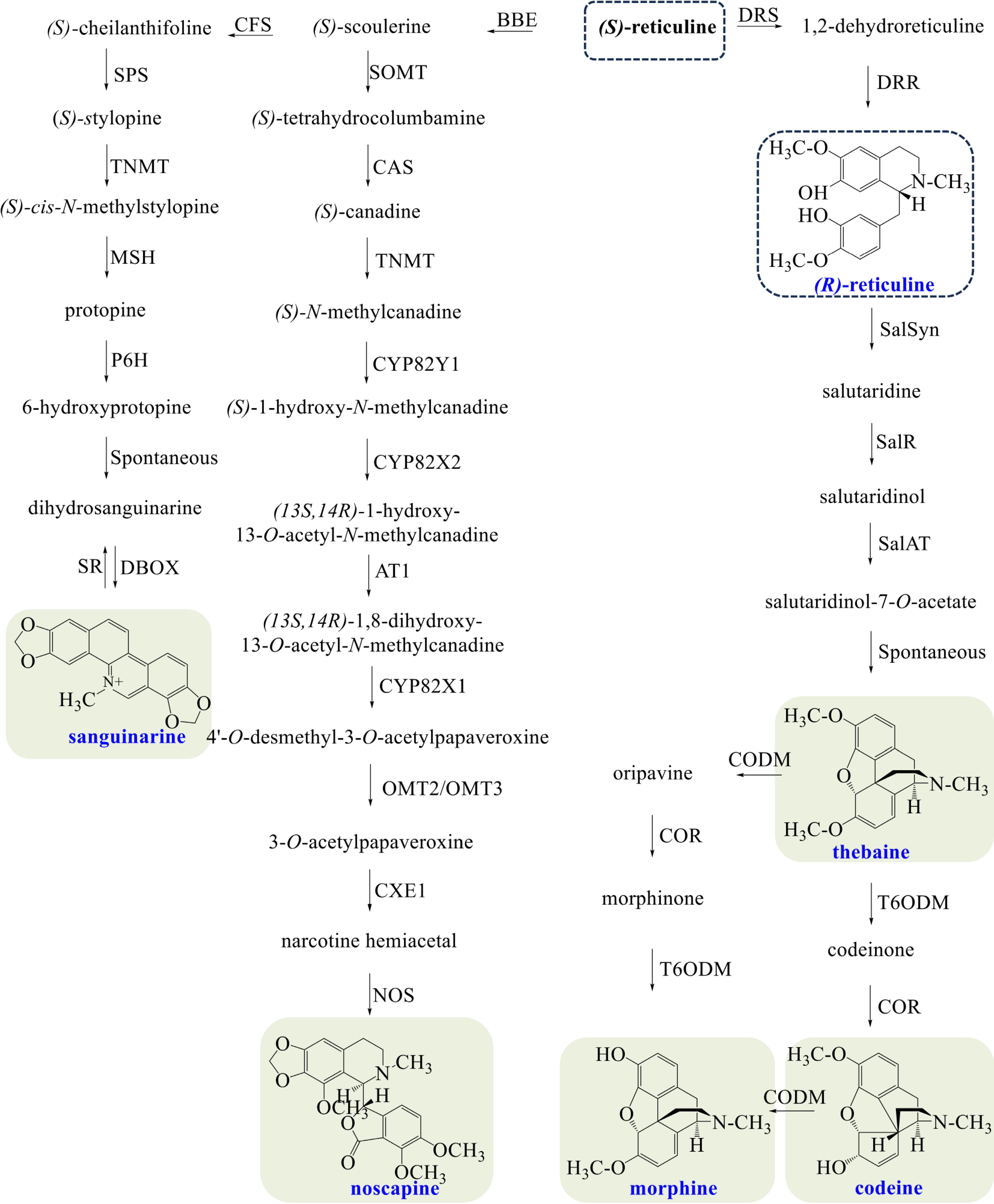
Figure 3.
Biosynthetic pathways of morphine, codeine, thebaine, noscapine, and sanguinarine in opium poppy. The broken arrows represent a hypothetical pathway that has not yet been substantiated. Abbreviations: BBE, berberine bridge enzyme; CFS, (S)-cheilanthifoline synthase; SPS, stylopine synthase; TNMT, tetrahydroprotoberberine cis-N-methyltransferase; MSH, (S)-cis-N-methylastylopine 14-hydroxylase; P6H, protopine 6-hydroxylase; DBOX, dihydrobenzophenanthridine oxidase; SR, sanguinarine reductase; SOMT, soulerine 9-O-methyltransferase; CAS, canadine synthase; CYP82Y1, N-methylcanadine 1-hydroxylase; CYP82X2 1-hydroxy-N-methylcanadine 13-O-hydroxylase; AT1, 1,13-dihydroxy-N-methylcanadine 13-O-acetyltransferase; CYP82X1, 1-hydroxy-13-O-acetyl-N-methylcanadine 8-hydroxylase; OMT2/OMT3, O-methyltransferases 2/O-methyltransferases 3; CXE1, 3-O-acetylpapaveroxine carboxylesterase 1; NOS, noscapine synthase; DRS, 1,2-dehydroreticuline synthase; DRR, 1,2-dehydroreticuline reductase; SalSyn, salutaridine synthase; SalR, salutaridine reductase; SalAT, salutaridinol 7-O-acetyltransferase; T6ODM, thebaine 6-O-demethylase; COR, codeinone reductase; CODM, codeine O-demethylase.
-
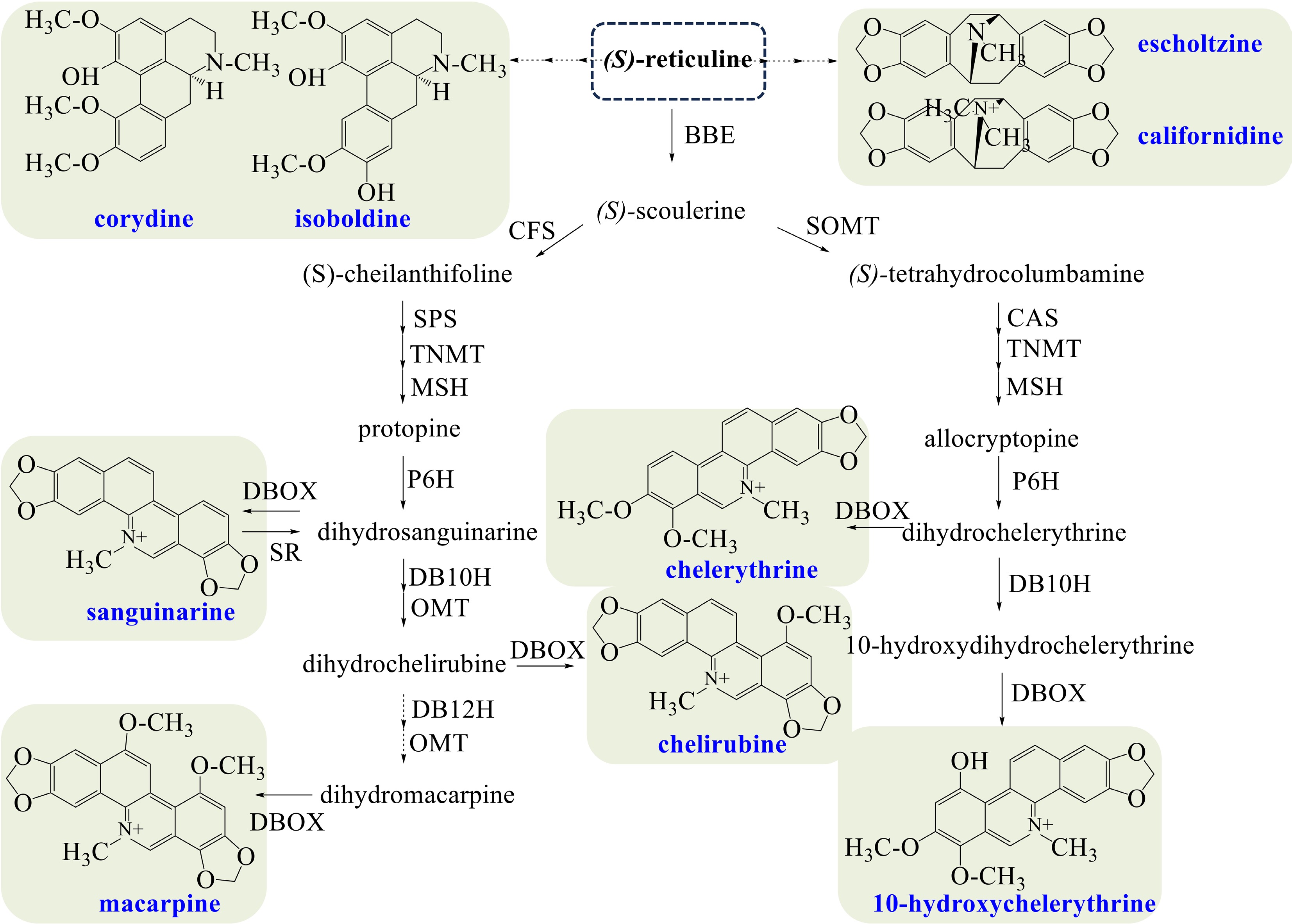
Figure 4.
Biosynthetic pathways of BIAs in California poppy. The broken arrows represent a hypothetical pathway that has not yet been substantiated. Abbreviations: BBE, berberine bridge enzyme; CFS, (S)-cheilanthifoline synthase; SPS, stylopine synthase; TNMT, tetrahydroprotoberberine cis-N-methyltransferase; MSH, (S)-cis-N-methylastylopine 14-hydroxylase; P6H, protopine 6-hydroxylase; DBOX, dihydrobenzophenanthridine oxidase; SR, sanguinarine reductase; DB10H, dihydrobenzophenanthridine alkaloid 10-hydroxylase; OMT, O-methyltransferase; DB12H, dihydrobenzophenanthridine alkaloid 12-hydroxylase; SOMT, soulerine 9-O-methyltransferase; CAS, canadine synthase.
-
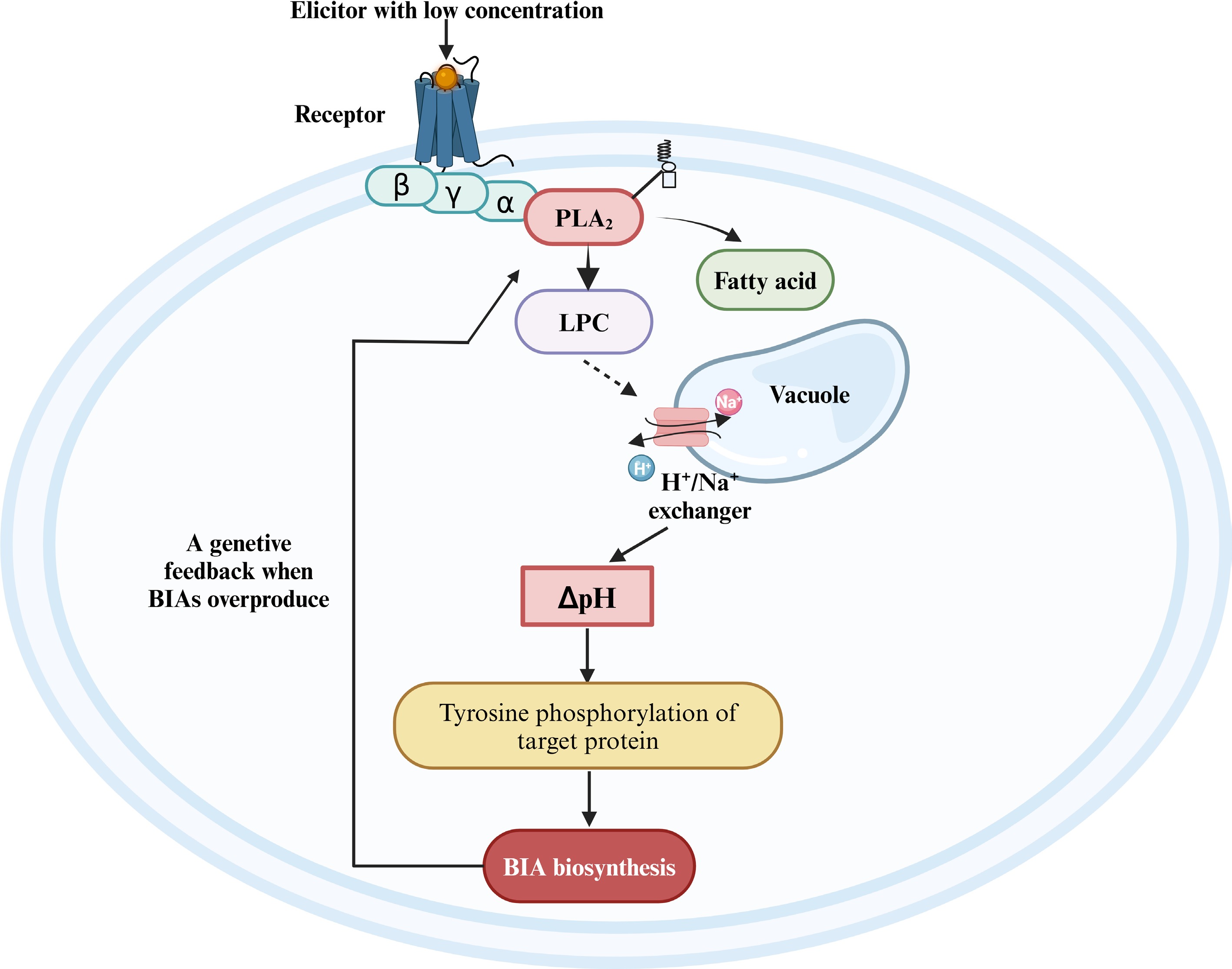
Figure 5.
The regulatory model of the jasmonate-independent pathway in the biosynthesis of BIAs. BIA, benzylisoquinoline alkaloid; PLA2, phospholipase A2; LPC, lysophosphatidylcholine. Reproduced with the permission from Ross et al.[74], Copyright 2006, Elsevier. This figure was created using the BioRender online tool (BioRender.com).
-
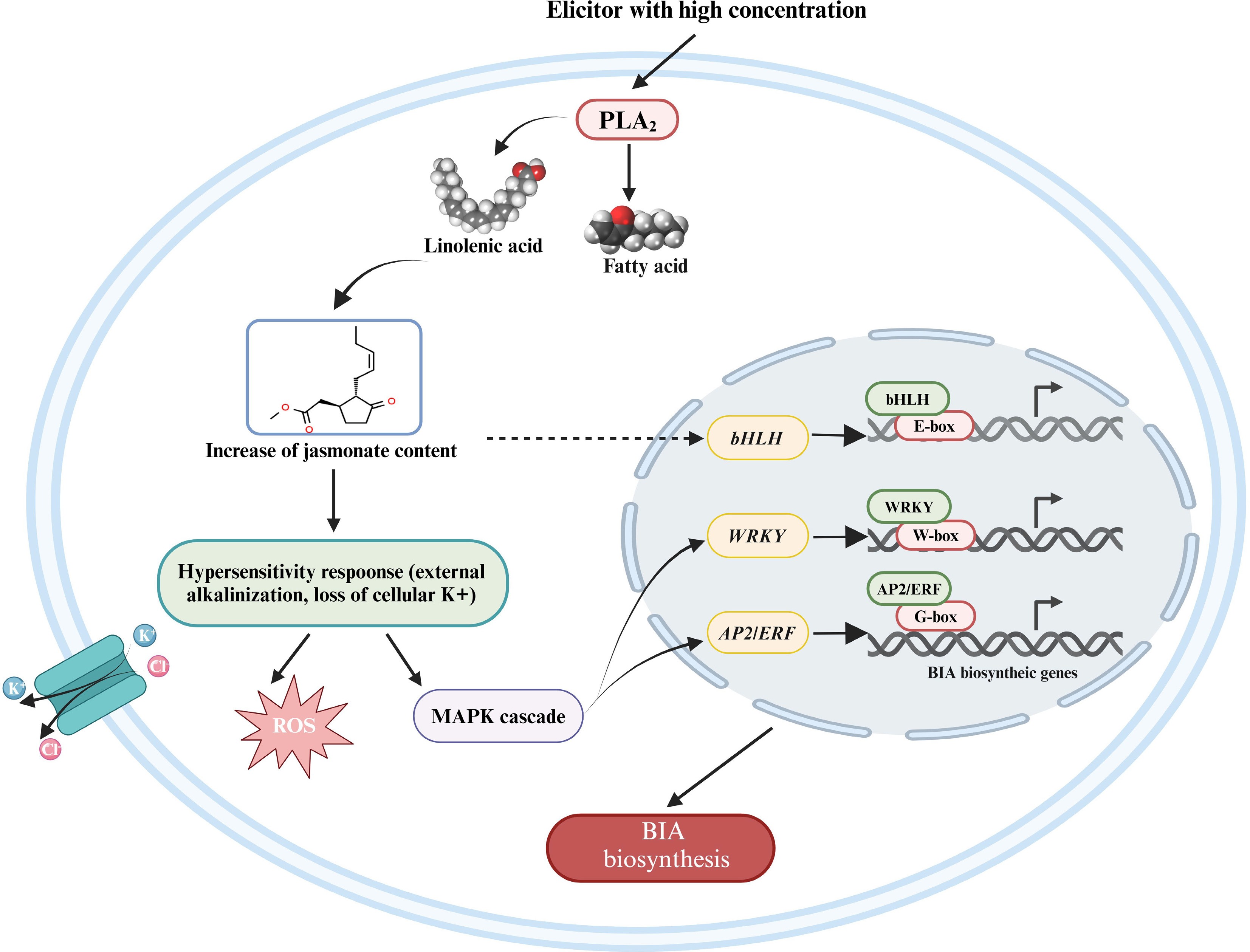
Figure 6.
The regulatory model of the jasmonate-dependent pathway in the biosynthesis of BIAs. A broken arrow represents a hypothesized signal cascade that has not yet been substantiated. BIA, benzylisoquinoline alkaloid; PLA2, phospholipase A2; ROS, reactive oxygen species; MAPK, mitogen-activated protein kinase. This figure was created using the BioRender online tool (BioRender.com).
-
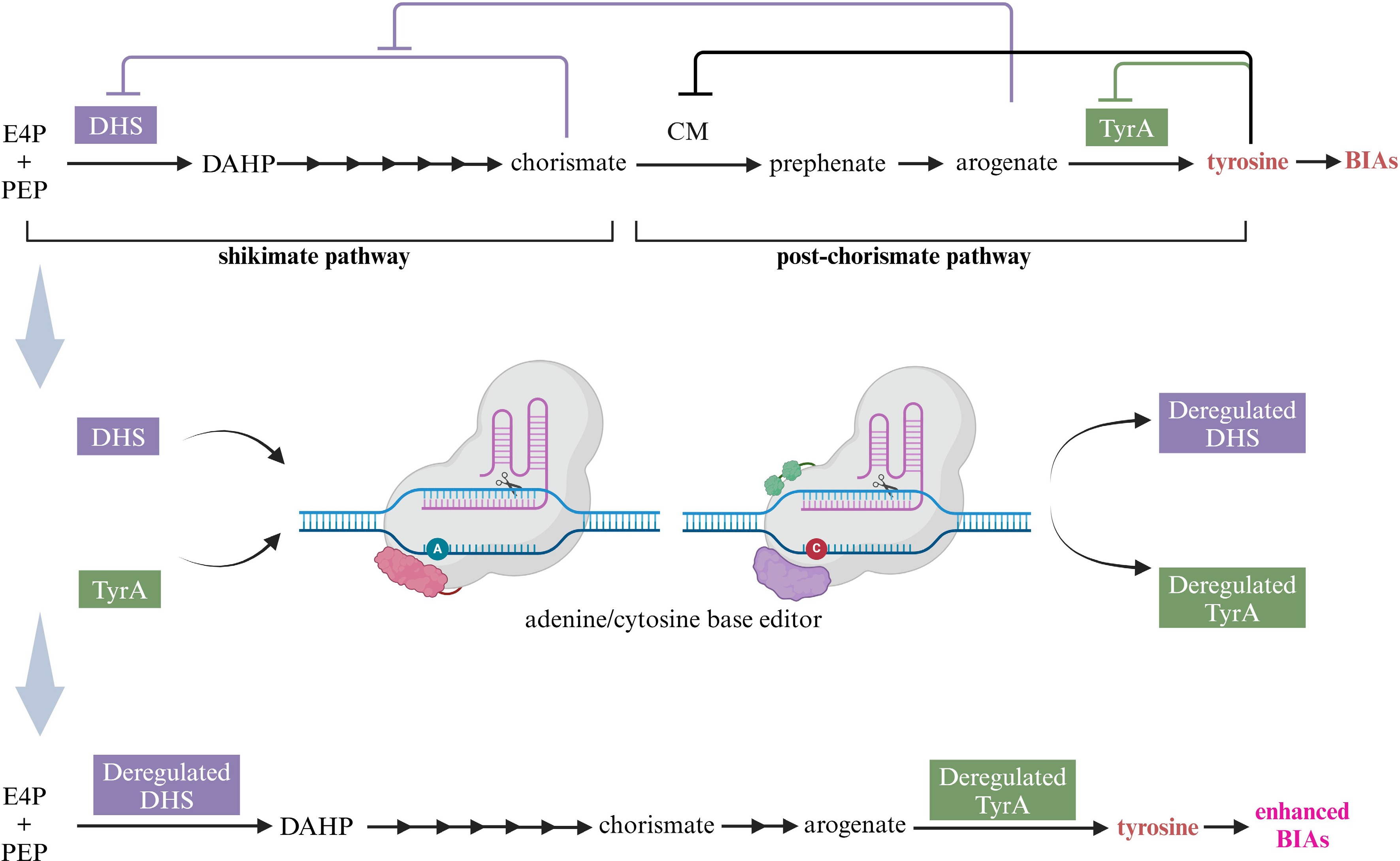
Figure 7.
Acquiring higher BIA yield through channeling more tyrosine via deregulated DHS and TyrA with the aid of base editing technology. E4P, erythrose 4-phosphate; PEP, phosphoenolpyruvate, DAHP, 3-deoxy-D-arabinoheptulosonate 7-phosphate; DHS, DAHP synthase; CM, chorismate mutase; TyrA, arogenate dehydrogenase; BIAs, benzylisoquinoline alkaloids. This figure was created using the BioRender online tool (BioRender.com).
-
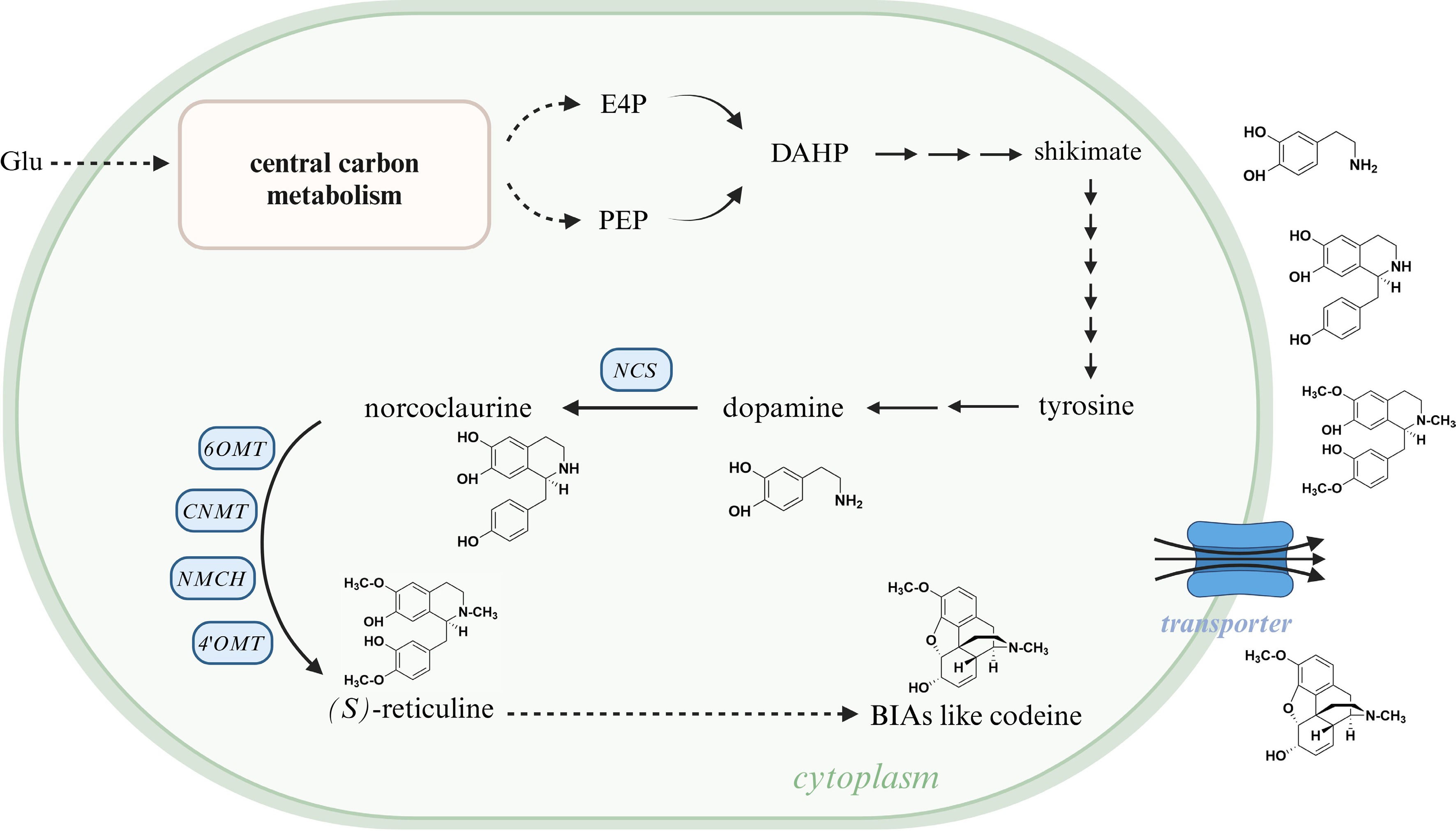
Figure 8.
Schematic of the function of heterologous transporters in BIA-producing microorganisms. The substrate takes glucose as an example; transported BIAs take dopamine, norcoclaurine, (S)-reticuline, and codeine as examples. Glu, glucose; E4P, erythrose 4-phosphate; PEP, phosphoenolpyruvate; DAHP, 3-deoxy-D-arabinoheptulosonate 7-phosphate; NCS, (S)-norcoclaurine synthase; 6OMT, 6-O-methyltransferase; CNMT, (S)-coclaurine N-methyltransferase; NMCH, N-methylcoclaurine 3'-hydroxylase; 4'OMT, 3'-hydroxy-N-methylcoclaurine 4'-O-methyltransferase; BIA, benzylisoquinoline alkaloid. This figure was created using the BioRender online tool (BioRender.com).
-
Plant Type Alkaloid Formula Property Ref. Coptis species Berberine Berberine C20H18NO4 Anti-inflammatory, anti-oxidant, anti-diabetic, neuro-protective, anti-cancer [27−30] Coptisine C19H14NO4 Anti-cancer, anti-inflammatory, anti-gastrointestinal [31] Epiberberine C20H18NO4 Anti-adipogenesis, anti-dyslipidemia, anti-cancer, anti-bacterial [32] Columbamine C20H20NO4 Suppresses cancer cells, anti-hypercholesterolemic [33,34] Palmatine C21H22NO4 Anti-Alzheimer's disease, anti-microbial, gastroprotective, hepatoprotective, anti-inflammatory, anti-cancer [35] Jatrorrhizine C20H20NO4 Anti-diabetic, anti-microbial and anti-protozoal, effects on the central nervous system, anti-cancer [12] Aporphine Magnoflorine C20H24NO4 Anti-diabetic, anti-inflammatory, neuropsychopharmacological, hypotensive, anti-fungal [11] Opium poppy Morphinan Morphine C17H19NO3 Analgesic [36] Morphinan Codeine C18H21NO3 Narcotic or opioid analgesic [37] Morphinan Thebaine C19H21NO3 Used for semi-synthesis of pain-relievers [37] Phthalideisoquinoline Noscapine C22H23NO7 Cough suppressant and anti-cancer [37] 1-benzylisoquinoline Papaverine C20H21NO4 Muscle relaxant [37] California poppy Benzophenanthridine Sanguinarine C20H14NO4 Anti-microbial [16] Chelirubine C21H16NO5 As a DNA fluorescent probe [38] Macarpine C22H18NO6 Interacting with DNA, against cancer cell lines [39] Chelerythrine C21H18NO4 Anti-bacterial, antineoplastic [40] Aporphine Corydine C20H23NO4 μ-opioid receptor agonist [41] Isoboldine C19H21NO4 Inhibition on the LPS-stimulated mRNA levels of IL-6 and IL-1β [42] Table 1.
Benzylisoquinoline alkaloids produced in Coptis species, opium poppy, and California poppy.
-
Species Origin Elicitor Duration Concentration Alkaloid Enhancement rate Ref. Opium poppy Biotic Botrytis sp. 80 h 1 mL/50 mL cell culture Sanguinarine 100 folds [77] Acinetobacter 72 h 10 mL Morphine 1044% [78] Noscapine 936% Papaverine 349% Kocuria sp. 72 h 10 ml Thebaine 718% [78] Acinetobacter sp. + Marmoricola sp. 2 h × 2 times 1 × 108 CFU mL−1 Morphine 2250% [79] Papaverine 36.4% Noscapine 53.3% Abiotic GA3+TRIA 90 d 10−6 M Morphine 60.3% [51] MeJA 12 h 100 mM Morphine 1.8 folds [80] Noscapine 1.6 folds Thebaine 3.2 folds Wound 5 h / Morphine / [81] Thebaine Wound / / Papaverine 125% [82] Narcotine 133% California poppy Biotic Yeast glycoprotein 24 h 1 μg·mL−1 Sanguinarine / [83,84] Chelirubine Macarpine 10-OHchelerythrine Abiotic MeJA 136 h 100 μM Sanguinarine / [85] 10-Hydroxychelerythrine Chelerythrine Chelirubine Macarpine Dihydrochelirubine MeJA + SA 48 h 0.5 mg + 0.02 mg·g−1 FCW Sanguinarine 980% [86] MeJA, methyl jasmonate; SA, salicylic acid; GA3, gibberellic acid; TRIA, triacontanol; /, data is not shown in reports. Table 2.
Positive impacts of abiotic and biotic elicitors on the BIA content in opium poppy and California poppy.
-
Plant source Transcription factors Target enzymes Enhanced alkaloids Ref. Coptis japonica/
Coptis chinensisCjWRKY1 All genes involved in berberine biosynthesis / [104] CjWRKY1 CYP80B2, 4'OMT and CYP719A1 / [105] CjWRKY1 EcCYP719A3, EcP6H, EcG3OMT, and EcG11OMT Sanguinarine, chelirubine, chelerythrine, allocryptopine, protopine, and 10-hydroxychelerythrine [106] CcWRKY7; CcWRKY29; CcWRKY32 CcCNMT protoberberine [107] CjbHLH1 All berberine biosynthetic enzyme genes (TYDC, NCS, 6OMT, CNMT, CYP80B2, 4'OMT, BBE, SMT and CYP719A1) / [103] CjbHLH1 4'OMT, CYP719A1 / [105] CcbHLH001; CcbHLH0002 CcBBE and CcCAS Five main BIA [108] 17 members of CcAP2/ERFs; family CcCAS, CcCTS, CcCoOMT, CcNMCH, and Cc4'OMT Berberine, columbamine, coptisine, epiberberine, jatrorrhizine, and palmatine [99] Opium poppy PsWRKY TYDC / [82] California poppy EcbHLH1-1; EcbHLH1-2 6OMT, 4'-OMT and CYP719A3 Sanguinarine [101] EcAP2/ERF2; EcAP2/ERF3; EcAP2/ERF4; EcAP2/ERF12 Ec6OMT and EcCYP719A5 / [109] Arabidopsis thaliana AtWRKY1 EcCYP80B1 and EcBBE BIAs in opium poppy and California poppy [110] Table 3.
Transcription factors targeting various BIA biosynthetic enzymes and alkaloids.
-
Host Produced BIAs Strategy Yield Ref. Escherichia coli (S)-reticuline De novo synthesis 46.0 mg·L−1 culture medium [131] (S)-reticuline De novo synthesis 384 μM [132] (S)-reticuline De novo synthesis 55 mg·L−1 within 1 h [133] Thebaine De novo synthesis 2.1 mg·L−1 [134] Saccharomyces cerevisiae (S)-reticuline De novo synthesis 80.6 μg·L-1 [135] Reticuline De novo synthesis 19.2 μg·L−1 [136] Magnoflorine De novo synthesis 75.8 mg·L−1 [137] Palmatine, berberine, chelerythrine, sanguinarine, and chelirubine De novo synthesis 38.1 mg·L−1 (chelerythrine) [40] Thebaine De novo synthesis 6.4 ± 0.3 µg·L−1 [138] Codeine, morphine, hydromorphone, hydrocodone, and oxycodone Synthesis from thebaine 131 mg·L−1 (total opioid titers) [139] Codeine and morphine Synthesis from (R)-reticuline / [140] Sanguinarine De novo synthesis 448.64 mg·L−1 [141] Sanguinarine De novo synthesis 1.8 mg·L−1 [142] Noscapine De novo synthesis ~2.2 mg·L−1 [143] Noscapine Biosynthesis from (S)-canadine 1.64 ± 0.38 μM [144] Tetrahydropapaverine De novo synthesis 121µg·L−1 [145] Combination cultures of
E. coli and S. cerevisiae cellsMagnoflorine and scoulerine De novo synthesis 7.2 and 8.3 mg·L−1 culture medium [133] /, data is not shown in reports. Table 4.
The rebuilt synthesis pathways of some BIAs in microbes.
Figures
(8)
Tables
(4)