-
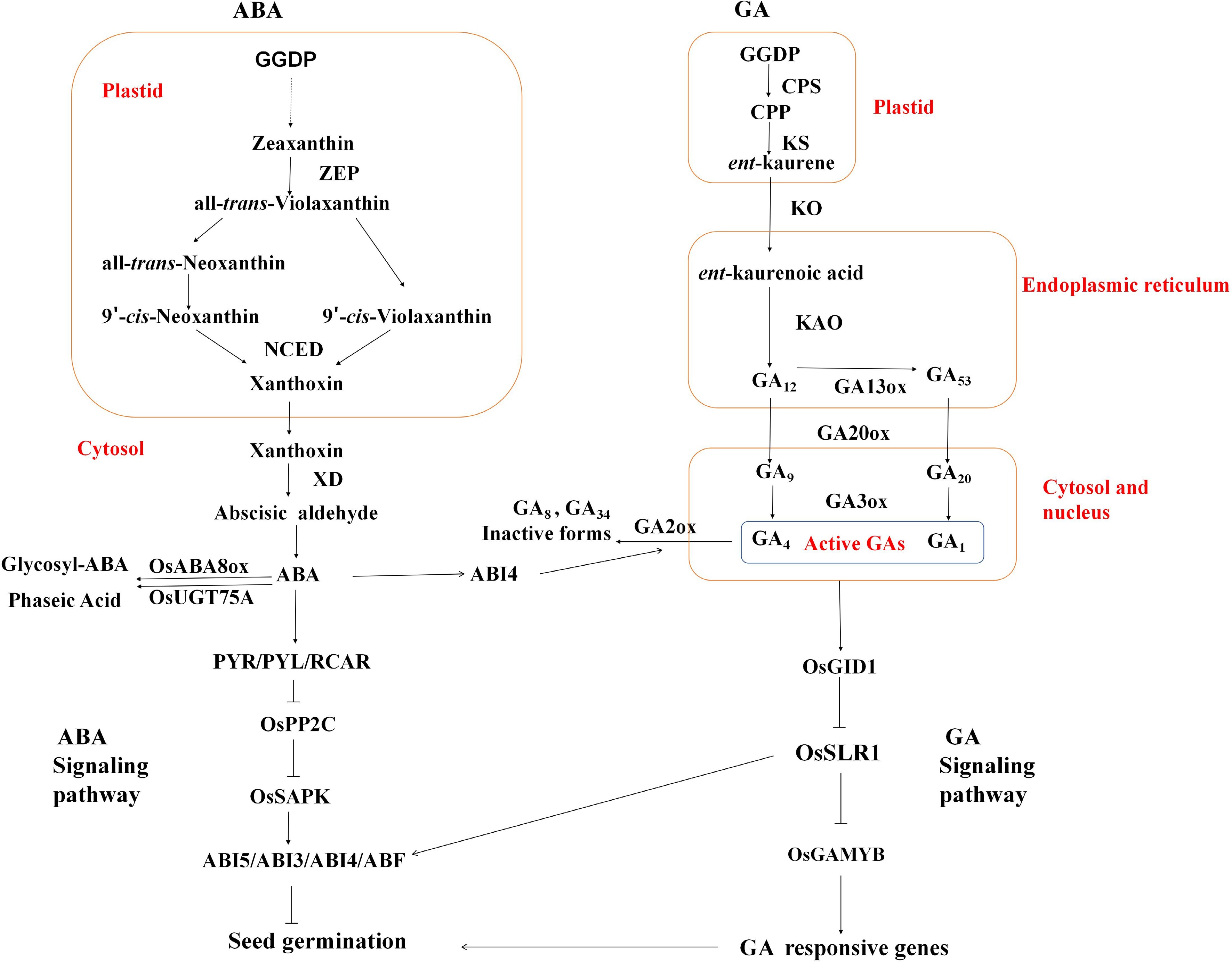
Figure 1.
The biosynthesis, metabolism, and signaling pathways of abscisic acid (ABA) and gibberellin (GA). GGDP: Geranylgeranyl diphosphate; ZEP: Zeaxanthin epoxidase; NCED: 9-cis-Epoxycarotenoid dioxygenase; XD: Xanthoxin dehydrogenase; CPS: ent-Copalyl diphosphate synthase; KS: ent-Kaurene synthase; CPP: ent-Copalyl diphosphate; KO: ent-Kaurene oxidase; KAO: ent-Kaurenoic acid oxidase.
-
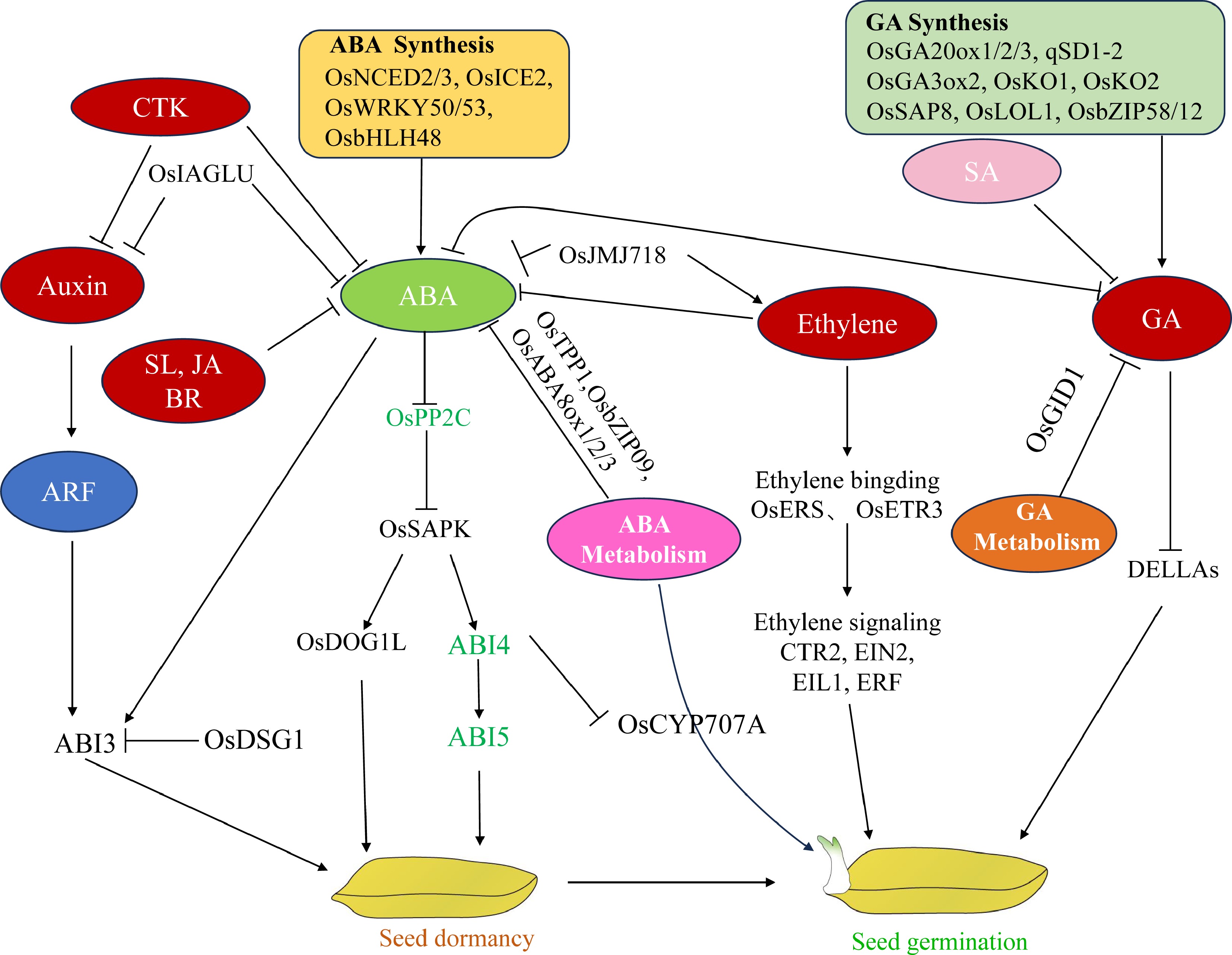
Figure 2.
Pattern diagram of plant hormone regulation of rice seed dormancy and germination.
-
Genes Gene accession number Signaling pathway Ref. OsNCED3 LOC_Os03g44380 Mutant seeds show a decreased ABA content and PHS. [16] OsbZIP10/ABI5/ OREB1 LOC_Os01g64000 Induced by ABA and high salinity, it negatively regulates rice seed germination. [18] OsDOG1L-3 LOC_Os01g20030 ABA positively regulates OsDOG1L-3 expression, and OsDOG1L-3 positively regulates the ABA pathway in a feedback manner, enhancing seed dormancy. [19] OsTRAB1 LOC_Os08g36790 Regulates ABA-induced transcriptional responses and plays an important role in regulating embryo maturation and dormancy. [18] SAPK2 LOC_Os07g42940 Mutants are insensitive to ABA during germination and post-germination stages. [20] OsFbx352 LOC_Os10g03850 OsFbx352 plays a role in the regulation of glucose-induced seed germination inhibition by targeting ABA metabolism. [21] OsWRKY29 LOC_Os07g02060 OsWRKY29 is a negative regulator of rice dormancy. The presence of ABA inhibits the expression of OsWRKY29, increasing the transcription of OsVP1 and OsABF1 and ultimately increasing rice seed dormancy. [22] OsWRKY50 LOC_Os11g02540 OsWRKY50 negatively regulates ABA-dependent seed germination by directly inhibiting the expression of the rice ABA biosynthesis gene OsNCED5. [23] OsWRKY53 LOC_Os05g272730 OsWRKY53 directly binds to the promoters of ABA catabolic genes OsABA8ox1 and OsABA8ox, inhibiting their expression, which results in ABA accumulation and further inhibits rice seed germination. [22] OsGAMYB LOC_Os01g0812000 MYBS1 and MYBGA are two MYB transcription factors. GA enhances the co-nuclear transport of MYBGA and MYBS1 and forms a stable binary MYB-DNA complex that activates α-amylase gene expression. Transcription factor 14 and OsGAmyb are downstream target genes of miR319. miR319 negatively regulates rice tillering and yield per plant by targeting OsTCP21 and OsGAmyb. [24] OsbHLH048 LOC_Os02g52190 bHLH transcription factors SD6 and ICE2 indirectly act on the ABA synthesis gene NCED2 through OsbHLH048 to regulate the ABA content in seeds and antagonistically regulate rice seed dormancy. [2] OsTPP1 LOC_Os02g44230 OsTPP1 controls rice seed germination through crosstalk with the ABA catabolic pathway. [25] Sdr4 LOC_Os07g39700 Positively regulated by OsVP1, mutants are insensitive to ABA. [26] SD6 LOC_Os06g06900 Negatively regulates dormancy and targets ABA8ox3 to indirectly regulate NCED2. [19] ICE2 LOC_Os01g70310 Positively regulates dormancy and antagonistically regulates ABA metabolism and synthesis with SD6. [2] ABA8ox3 LOC_Os09g28390 An ABA catabolic gene directly regulated by SD6/ICE2. [2] NCED2 LOC_Os12g24800 An ABA synthesis gene indirectly regulated by SD6/ICE2 through OsbHLH048. [2] DG1 LOC_Os03g12790 A MATE transporter that regulates the long-distance transport of ABA from leaves to caryopses and responds to temperature changes. [27] OsSAE1 LOC_Os06g43220 Directly binds to the promoter of the key ABA signaling pathway gene OsABI5, inhibiting its expression and promoting rice seed germination. [28] OsG LOC_Os03g01014 G affects seed dormancy through the interaction with NCED3 and psy and then regulates ABA synthesis [29] Table 1.
Genes regulating rice seed germination and dormancy through abscisic acid (ABA) metabolism, transport, and signal transduction.
-
Genes Gene accession number Signaling pathway Ref. IPA1 LOC_Os08g39890 IPA1 mediates part of the effect of miR156 on seed dormancy by directly regulating many genes in the GA pathway. [52] SAPK10 LOC_Os03g41460 SAPK10 increases the endogenous GA level and inhibits seed germination. [53] OsbZIP12 LOC_Os01g64730 OsbZIP12 inhibits GA synthesis and regulates rice seed germination. [53] WRKY72 LOC_Os11g29870 WRKY72 directly targets LRK1, activates its transcription, and inhibits OsKO2 (ent-kaurene oxidase), thereby reducing the endogenous GA level and inhibiting rice seed germination. [54] OsGA20ox2 LOC_Os01g66100 OsGA20ox2 is expressed in developing seeds, including endosperm tissues, and regulates rice seed germination through the GA signaling pathway. [55] OsLOL1 LOC_Os08g06280 OsLOL1 increases the GA level, affects the aleurone layer programmed cell death (PCD) process, and promotes rice seed germination. [56] OsSLR1 LOC_Os03g49990 GA relieves the inhibition of OsUDT1/OsTDR by OsSLR1, thereby positively regulating rice fertility. [22] OsKO1 LOC_Os06g37330 The mutation of OsKO1 reduces enzyme activity, decreasing the contents of GA1, GA3, and GA53 and delaying seed germination. [57] qSD1-2 LOC_Os01g66100 Involved in GA biosynthesis. [55] OsFIE1 LOC_Os08g04290 Osfie1 inhibits seed germination and aleurone layer thickening by inhibiting GA accumulation in rice endosperm [58] Table 2.
Genes regulating rice seed germination and dormancy through gibberellin (GA) metabolism, transport, and signal transduction.
Figures
(2)
Tables
(2)