-
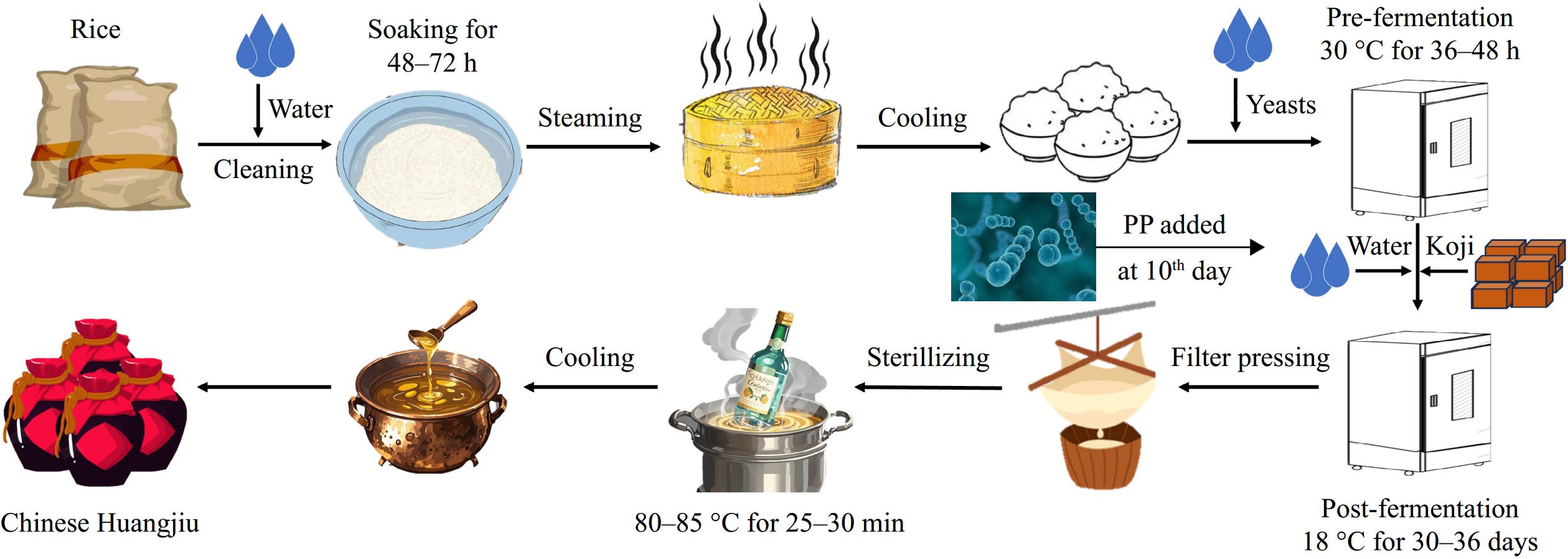
Figure 1.
Huangjiu fermentation process.
-
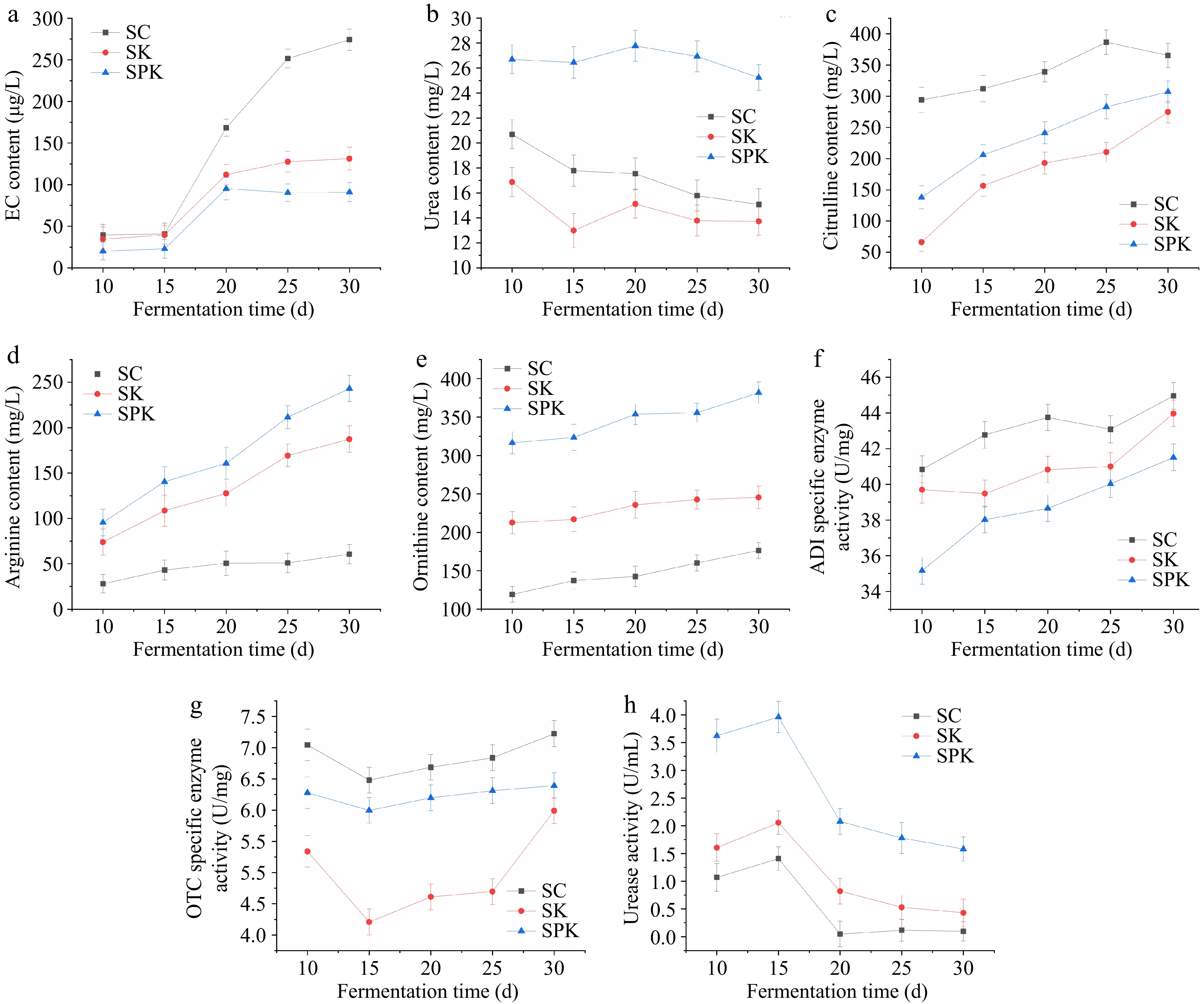
Figure 2.
Variation in EC, arginine-related metabolites and enzymes in the processing of Huangjiu under different fermentation systems. (a) Effect of different fermentation systems on EC formation. (b) Urea content. (c) Citrulline content. (d) Arginine content. (e) Ornithine content. (f) ADI-specific activity. (g) OTC-specific activity. (h) Urease activity. SPK, BTN2 knockout strain with PP; SC, S. cerevisiae; SK, BTN2 knockout strain.
-
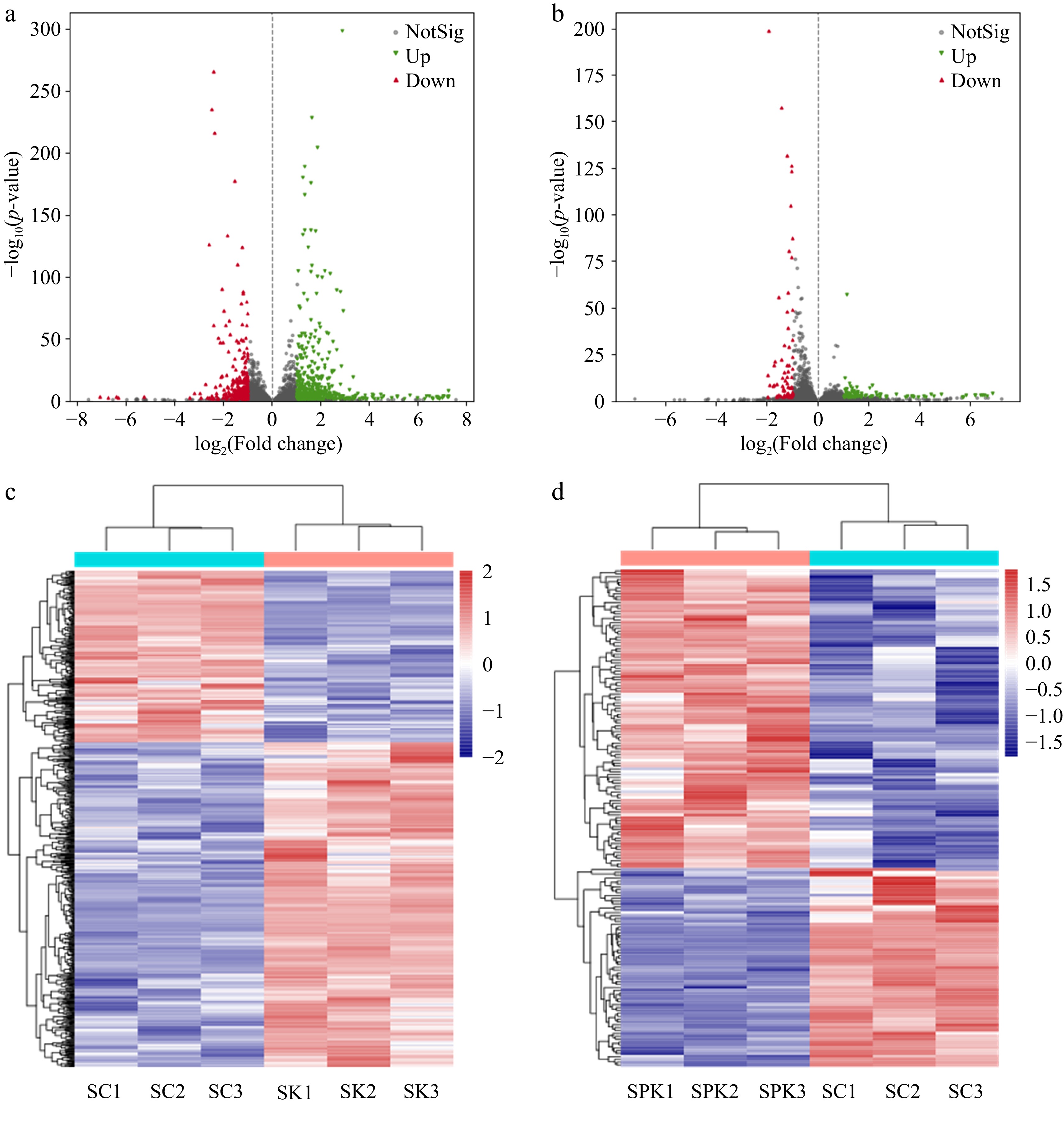
Figure 3.
Heatmap and volcano plot of DEGs. (a) DEGs in SK. (b) DEGs in SPK. (c) Volcano plot of DEGs in SC and SK. (d) Volcano plot of DEGs in SC and SPK. SPK, BTN2 knockout strain with PP; SK, BTN2 knockout strain.
-
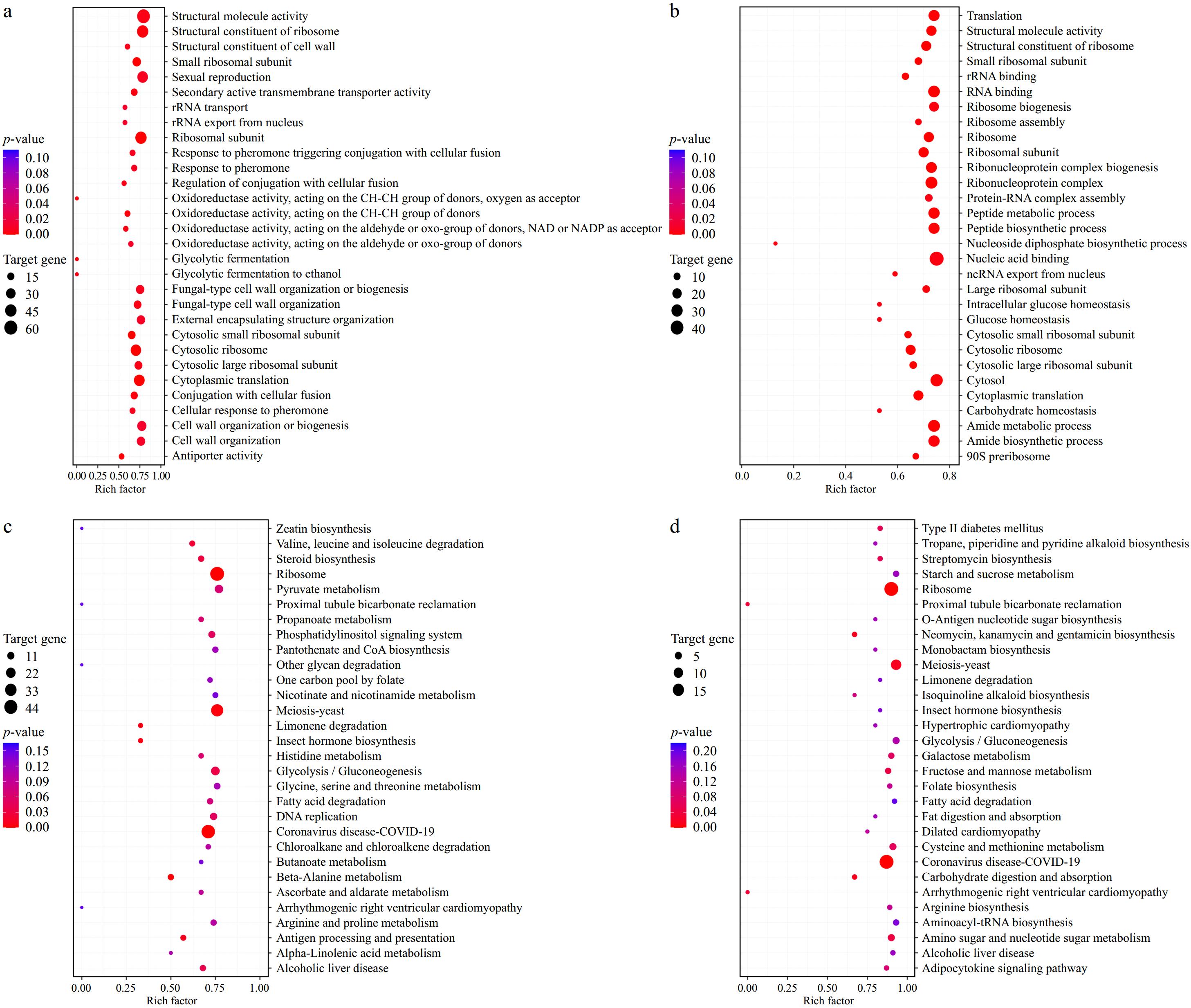
Figure 4.
GO and KEGG analysis. (a) GO analysis of SK. (b) GO analysis of SPK. (c) KEGG analysis of SK. (d) KEGG analysis of SPK. SPK, BTN2 knockout strain with PP; SK, BTN2 knockout strain.
-

Figure 5.
The result of verification by RT-qPCR.
-
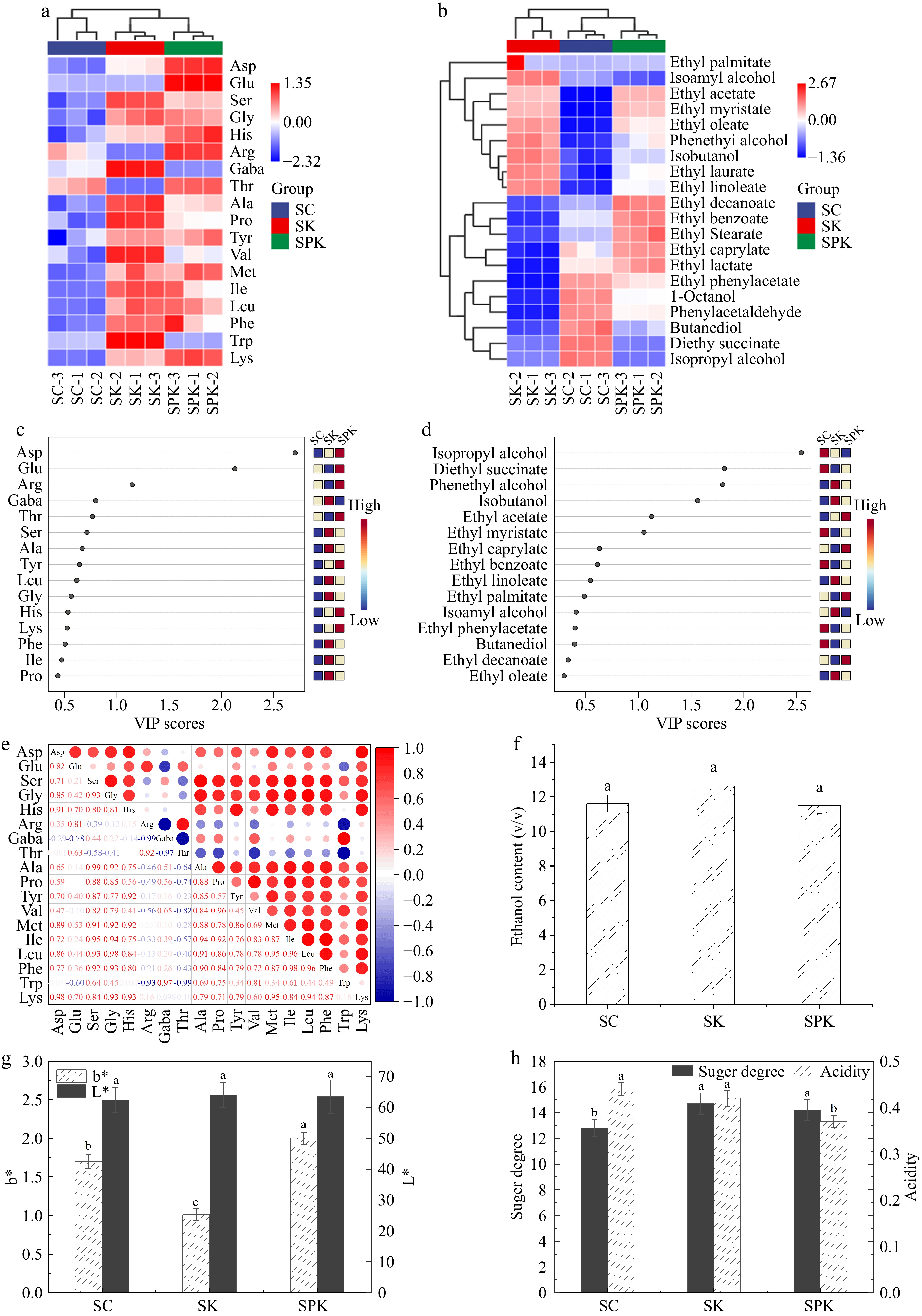
Figure 6.
Analysis of amino acids and volatile flavor substances in Huangjiu under different fermentation systems. (a) Cluster heat map analysis of free amino acids in Huangjiu. (b) variable importance inprojection (VIP) scores of free amino acids under different fermentation systems. (c) Heat map of free amino acid correlations. (d) Cluster heat map analysis of volatile flavor substances (e) VIP score of volatile flavor substances under different fermentation systems. (f) Alcohol content. (g) b* and L* values. (H) Sugar–acid ratio. SPK, BTN2 knockout strain with PP; SC, S. cerevisiae; SK, BTN2 knockout strain.
-
Genes Function Log2(Fold change) DEGs and their functions in SK Ard1 Predicted to enable DNA-binding transcription factor activity and RNA Polymerase II cis-regulatory region sequence-specific DNA-binding activity. 3.7122 Ntc20 Predicted to enable reduced nicotinamide adenine dinucleotide (NADH) dehydrogenase activity. Involved in NADH oxidation and glycolytic fermentation to ethanol. 3.563 Shq1 Involved in the cellular response to amino acid stimuli and transcription factor catabolic processes. Located in the inner nuclear membrane 3.3911 Erv15 Enables organic acid transmembrane transporter activity. Involved in organic acid transport. 3.3432 Mum3 Enables protein kinase activity. Involved in several processes 3.3293 Mga1 Predicted to enable DNA-binding transcription factor activity and sequence-specific DNA-binding activity. 2.909 Pex21 Enables adenosine triphosphate synthase (ATPase) activator activity and unfolded protein binding activity. 2.6193 Mud1 Enables mannosyltransferase activity. Involved in glycosylphosphatidylinisotol (GPI) anchor biosynthetic process. 2.4528 Cab1 Enables adenosine triphosphate (ATP)-dependent peptidase activity. 2.3866 Rrg8 Predicted to enable amino acid transmembrane transporter activity. 2.285 Asi2 Enables DNA-binding activity, bending and centromeric DNA-binding activity. 2.2283 Kip2 Involved in DNA repair. Located in the cytosol and nucleus. 2.1517 Rps9 Enables cyclin-dependent protein serine/threonine kinase regulator activity 2.1481 Gzf3 Enables cyclin-dependent protein serine/threonine kinase regulator activity. 2.1396 Kar1 Enables 3′–5′ DNA helicase activity and enzyme activator activity. 2.064 Yar1 Enables protein kinase inhibitor activity. Involved in fungal-type cell wall organization and negative regulation of protein kinase activity. 2.0084 Mak16 Predicted to enable oxidoreductase activity. Involved in protein targeting to membrane and protein transport into membrane rafts. 1.9673 Gar1 Predicted to enable DNA-binding activity. Involved in regulation of fungal-type cell wall biogenesis and regulation of the mitotic cell cycle. 1.9195 Osw1 Enables dicarboxylic acid transmembrane transporter activity and sulfate transmembrane transporter activity. −2.9978 Aro80 Enables protein transporter activity and unfolded protein binding activity. −2.4876 Rec104 Enables DNA secondary structure binding activity. −2.2241 Shy1 Enables cyclin-dependent protein serine/threonine kinase regulator activity. −1.5477 Bud27 Enables protein kinase activity. Involved in several processes, including DNA recombination, positive regulation of DNA-dependent DNA replication and protein phosphorylation. −1.2341 Lif1 Enables DNA replication origin binding activity and adenylate kinase activity. −1.2149 Sip4 Enables sequence-specific DNA-binding activity. −1.2094 Lam5 Enables DNA-binding transcription factor activity, RNA Polymerase II-specific activity and RNA Polymerase II cis-regulatory region sequence-specific DNA-binding activity −1.1971 Pib2 Involved in DNA recombinase assembly and maintenance of rDNA. −1.1861 Atg5 Enables RNA Polymerase I general transcription initiation factor activity. −1.165 Apl5 Enables sequence-specific DNA-binding activity. −1.1553 Vac17 Enables DNA-binding transcription factor activity. Involved in the response to xenobiotic stimulus. −1.1399 Fet3 Involved in positive regulation of transcription by RNA Polymerase II. −1.1133 Lre1 Enables DNA-binding transcription activator activity, RNA Polymerase II-specific. −1.046 DEGs and their functions in SPK Sbh1 Enables guanyl nucleotide exchange factor activity and protein transmembrane transporter activity. Contributes to protein-transporting ATPase activity. 3.0518 Utp4 Involved in maturation of small subunit (SSU) rRNA from tricistronic rRNA transcripts and positive regulation of transcription by RNA Polymerase I. 2.6671 Dpm1 Enables dolichyl-phosphate beta-D-mannosyltransferase activity. 1.5603 Gar1 Enables Box H/ACA small nucleolar (snoRNA) binding activity. 1.5532 Tif11 Enables RNA binding activity, ribosomal small subunit binding activity and translation initiation factor binding activity. 1.5054 Adk1 Enables DNA replication origin binding activity and adenylate kinase activity. 1.2293 Git1 Enables glycerol-3-phosphate transmembrane transporter activity and glycerophosphodiester transmembrane transporter activity. 1.2141 Dic1 Enables dicarboxylic acid transmembrane transporter activity. 1.1481 Egt2 Predicted to enable cellulase activity. 1.1403 Mcm4 Enables DNA replication origin binding activity and single-stranded DNA-binding activity. 1.129 Hxt6 Enables glucose transmembrane transporter activity and pentose transmembrane transporter activity. 1.1233 Mot3 Enables DNA-binding transcription factor activity 1.0971 Cin5 Enables DNA-binding transcription factor binding activity and sequence-specific DNA-binding activity. 1.0629 Kti12 Enables chromatin binding activity. 1.0457 Hop1 Enables four-way junction DNA-binding activity. −1.0204 Tah11 Enables DNA replication origin binding activity. −1.0225 Cbf2 Enables DNA-binding activity, bending and centromeric DNA-binding activity. −1.0359 Bur2 Enables cyclin-dependent protein serine/threonine kinase regulator activity. −1.0365 Lpx1 Enables triglyceride lipase activity. Involved in the triglyceride catabolic process. −1.054 Stn1 Enables single-stranded telomeric DNA-binding activity and translation elongation factor binding activity. −1.0597 Pcl10 Enables cyclin-dependent protein serine/threonine kinase regulator activity. −1.1792 Ast1 Predicted to enable oxidoreductase activity. Involved in protein targeting to the membrane and protein transport into membrane rafts. −1.1986 Rog3 Enables ubiquitin protein ligase binding activity. −1.2246 Ctf3 Involved in the initiation of DNA replication, establishment of mitotic sister chromatid cohesion and mitotic spindle assembly checkpoint signaling. −1.2298 Pga2 Involved in protein transport. Located in the endoplasmic reticulum and the nuclear envelope. −1.2993 Faa4 Enables long-chain fatty acid–coenzyme A (CoA) ligase activity. Involved in long-chain fatty acid import into the cell −1.4328 Tda6 Predicted to be involved in protein transport. Located in the cell periphery and fungal-type vacuoles −1.5558 Cyk3 Enables enzyme regulator activity. Involved in secondary cell septum biogenesis. −1.7224 Sps1 Enables protein kinase activity. Involved in several processes, including ascospore wall assembly and meiotic spindle disassembly −1.9151 Table 1.
DEGs in S. cerevisiae and their functions.
Figures
(6)
Tables
(1)