-
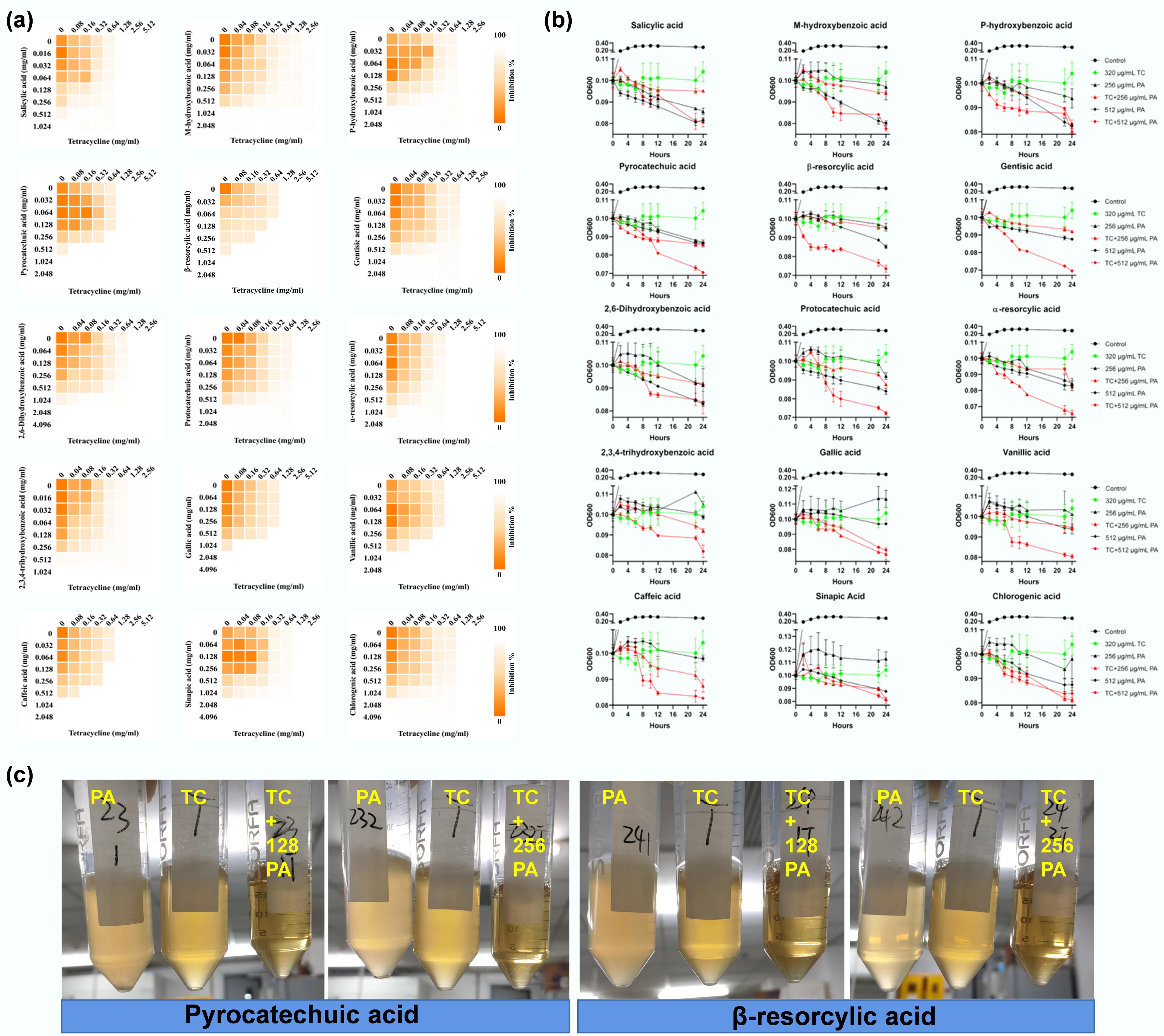
Figure 1.
Synergistic effects between phenolic acids and tetracycline. (a) Checkerboard assay revealing synergistic effects between phenolic acids and tetracycline against multidrug-resistant E. coli MG1655/RP4. Heatmap intensity reflects bacterial density. Data represent the averages of three biological replicates. (b) Time-dependent killing of MDR E. coli MG1655/RP4 by the combination of phenolic acids (PA) with tetracycline. (c) Combinations of tetracycline (320 µg mL−1) with two representative PAs, pyrocatechuic acid and β-resorcylic acid, at 128 and 256 µg mL−1 induce cell lysis.
-
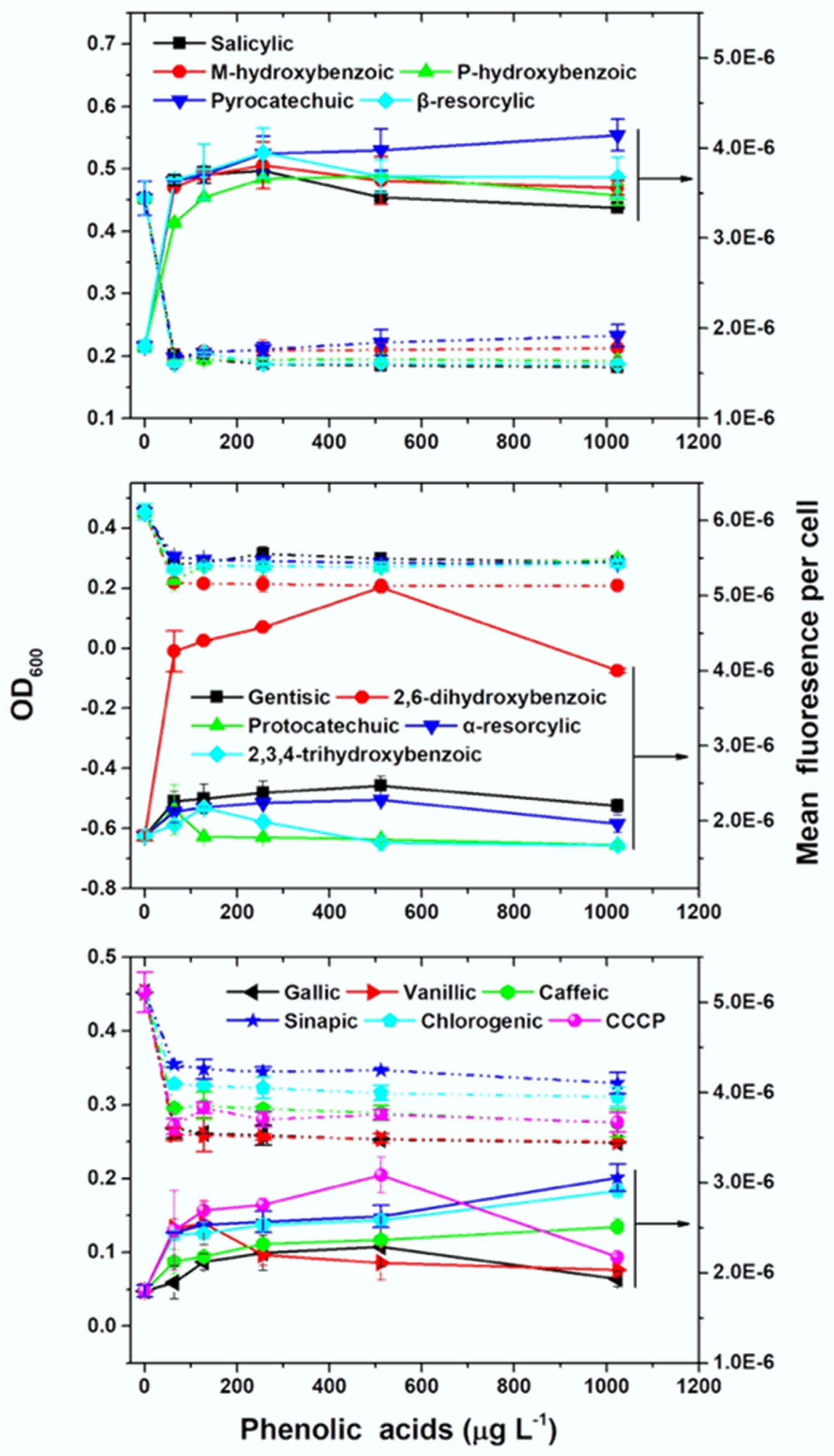
Figure 2.
Phenolic acids increase tetracycline uptake and inhibit the growth of E. coli MC4100/pTGM. The effects of 15 phenolic acids were tested alongside the efflux pump inhibitor carbonyl cyanide 3-chlorophenylhydrazone (CCCP).
-
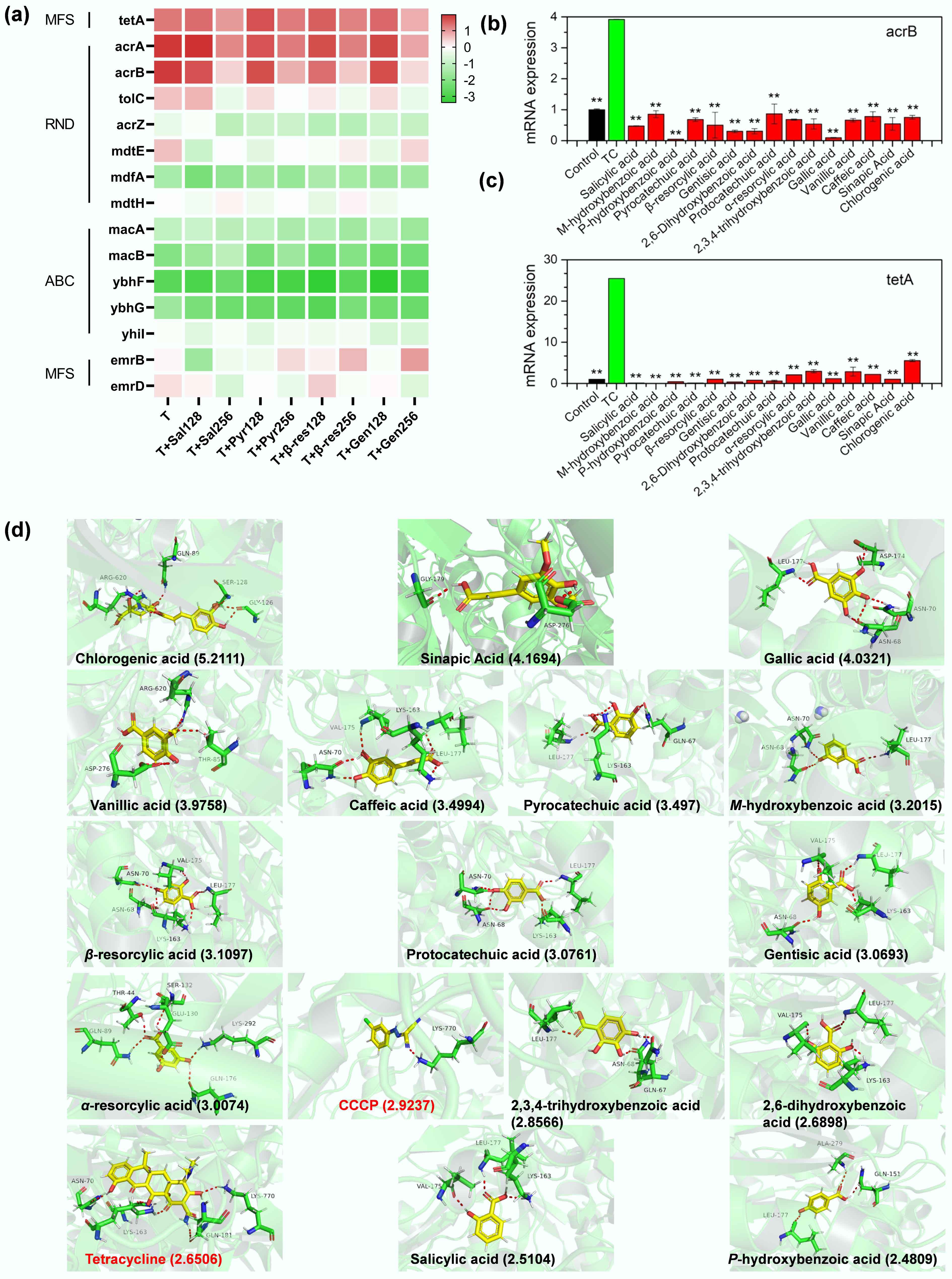
Figure 3.
Phenolic acids impair bacterial efflux pumps. (a) Transcriptomic analysis reveals dose-dependent downregulation of efflux pump genes by phenolic acids (128 and 256 μg mL−1; Sal: salicylic acid; Pyr: pyrocatechuic acid; β-res: β-resorcylic acid; Gen: gentistic acid) compared to tetracycline (T) alone. (b), (c) Phenolic acids decrease the expression of efflux pump genes acrB and tetA in MDR E. coli MG1655/RP4. The asterisk indicates a statistically significant difference compared with tetracycline (TC) alone. * p < 0.05, ** p < 0.01. (d) Molecular docking determines interactions between phenolic acids and the acrB protein, with total binding scores (in parentheses). Red dashed lines represent hydrogen bonds with labeled amino acid residues.
-
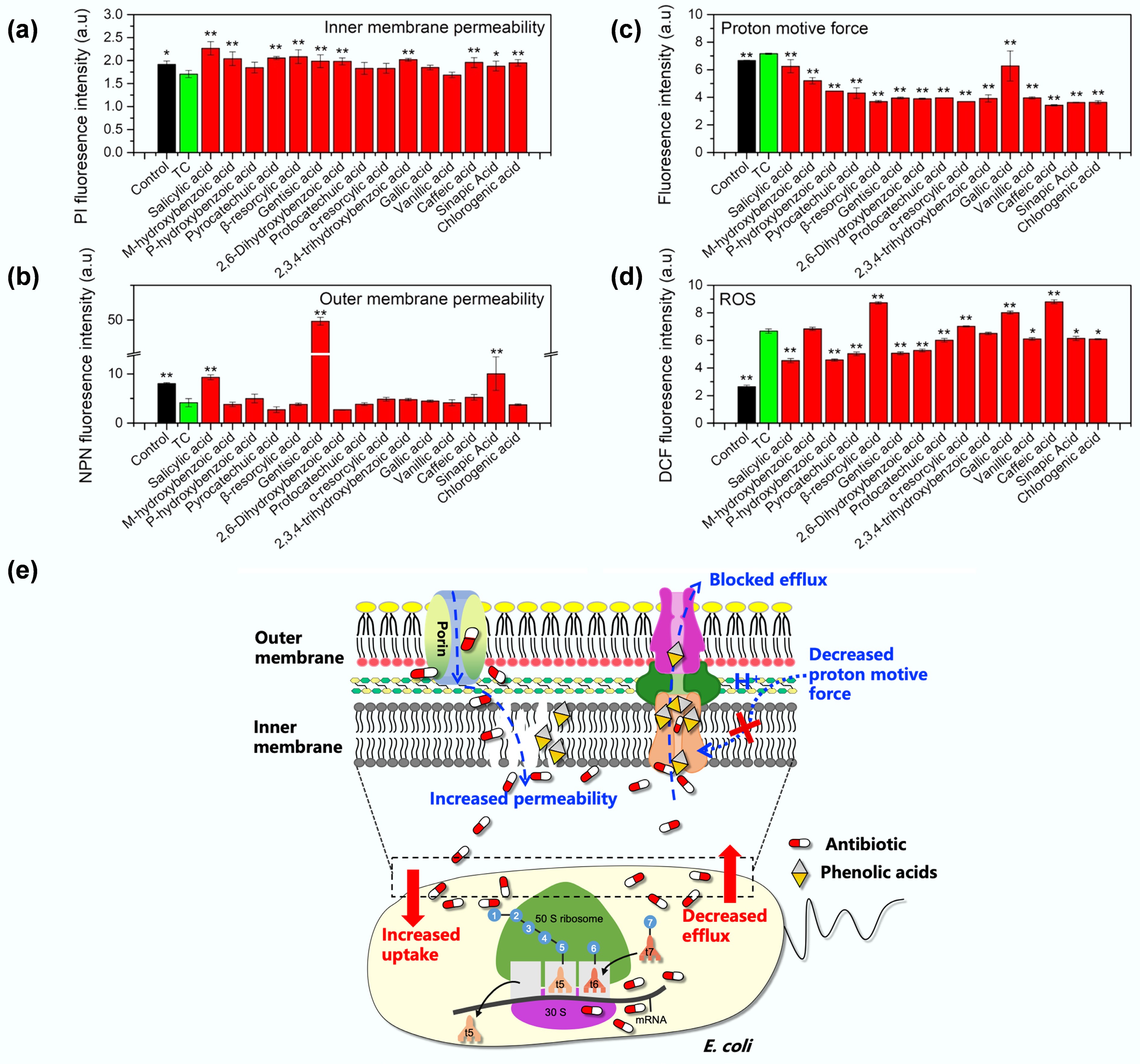
Figure 4.
Mechanisms underlying the synergetic effect of tetracycline and phenolic acids. Comparisons of (a) inner membrane permeability, (b) outer membrane permeability, (c) proton motive force, (d) reactive oxygen species (ROS) in MDR E. coli MG1655/RP4 upon exposure to tetracycline (TC) alone or phenolic acid-tetracycline combinations. Controls represent bacteria grown in LB medium without treatment. (e) Proposed mechanisms underlying the synergetic effect of tetracycline and phenolic acids: phenolic acids enhance tetracycline uptake by increasing inner membrane permeability and impairing efflux pump activity (either through direct binding to block the efflux pump or by depleting the energy source of proton motive force required for efflux) to reduce antibiotic efflux.
-
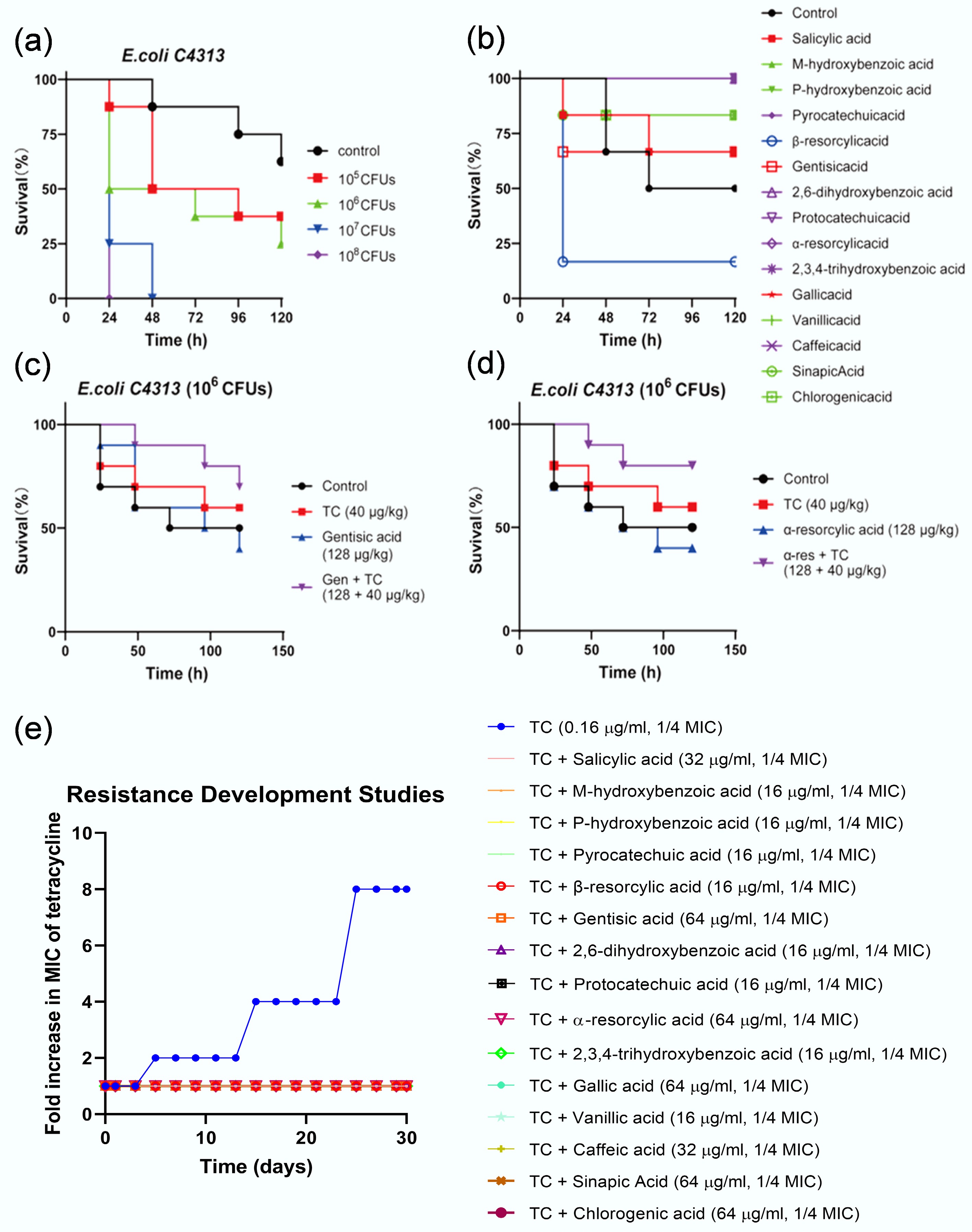
Figure 5.
Application potential evaluation of phenolic acid-tetracycline combinations. (a)–(d) Phenolic acid-tetracycline combinations effectively treat bacterial infections in Galleria mellonella. (a) Optimization of E. coli C4313 inoculum concentration for the G. mellonella infection assay. (b) Toxicity assessment of 15 phenolic acids in G. mellonella. (c), (d) Two representative phenolic acids of gentisic acid and resorcylic acid potentiate tetracycline potency in vivo. (e) Combinations of phenolic acids (1/4 MIC) with tetracycline (1/4 MIC; 0.16 µg mL−1) delay the emergence of tetracycline resistance in E. coli ATCC 25922.
-
Compound E. coli C4313 E. coli C4313∆acrB FIC Classfication Potentitation fold FIC Classfication Potentitation fold Salicylic acid 0.250 Synergy 16 0.281 Synergy 32 M-hydroxybenzoic acid 0.156 Synergy 16 0.281 Synergy 32 P-hydroxybenzoicacid 0.156 Synergy 16 0.281 Synergy 32 Pyrocatechuicacid 0.250 Synergy 8 0.281 Synergy 32 β-resorcylicacid 0.250 Synergy 16 0.500 Synergy 4 Gentisic acid 0.156 Synergy 16 0.500 Synergy 4 2,6-dihydroxybenzoic acid 0.156 Synergy 16 0.500 Synergy 4 Protocatechuicacid 0.250 Synergy 8 0.625 Independent 8 α-resorcylic acid 0.188 Synergy 16 0.750 Independent 4 2,3,4-trihydroxybenzoic acid 0.156 Synergy 8 0.750 Independent 4 Gallic acid 0.156 Synergy 16 0.750 Independent 4 Vanillic acid 0.156 Synergy 8 0.625 Independent 8 Caffeic acid 0.156 Synergy 16 0.625 Independent 8 Sinapic Acid 0.188 Synergy 8 0.750 Independent 4 Chlorogenic acid 0.156 Synergy 8 0.750 Independent 4 Table 1.
The fractional inhibitory concentration (FIC) values for combinations of phenolic acids and tetracycline against E. coli C4313 and its acrB-deleted mutant, E. coli C4313∆acrB
Figures
(5)
Tables
(1)