-
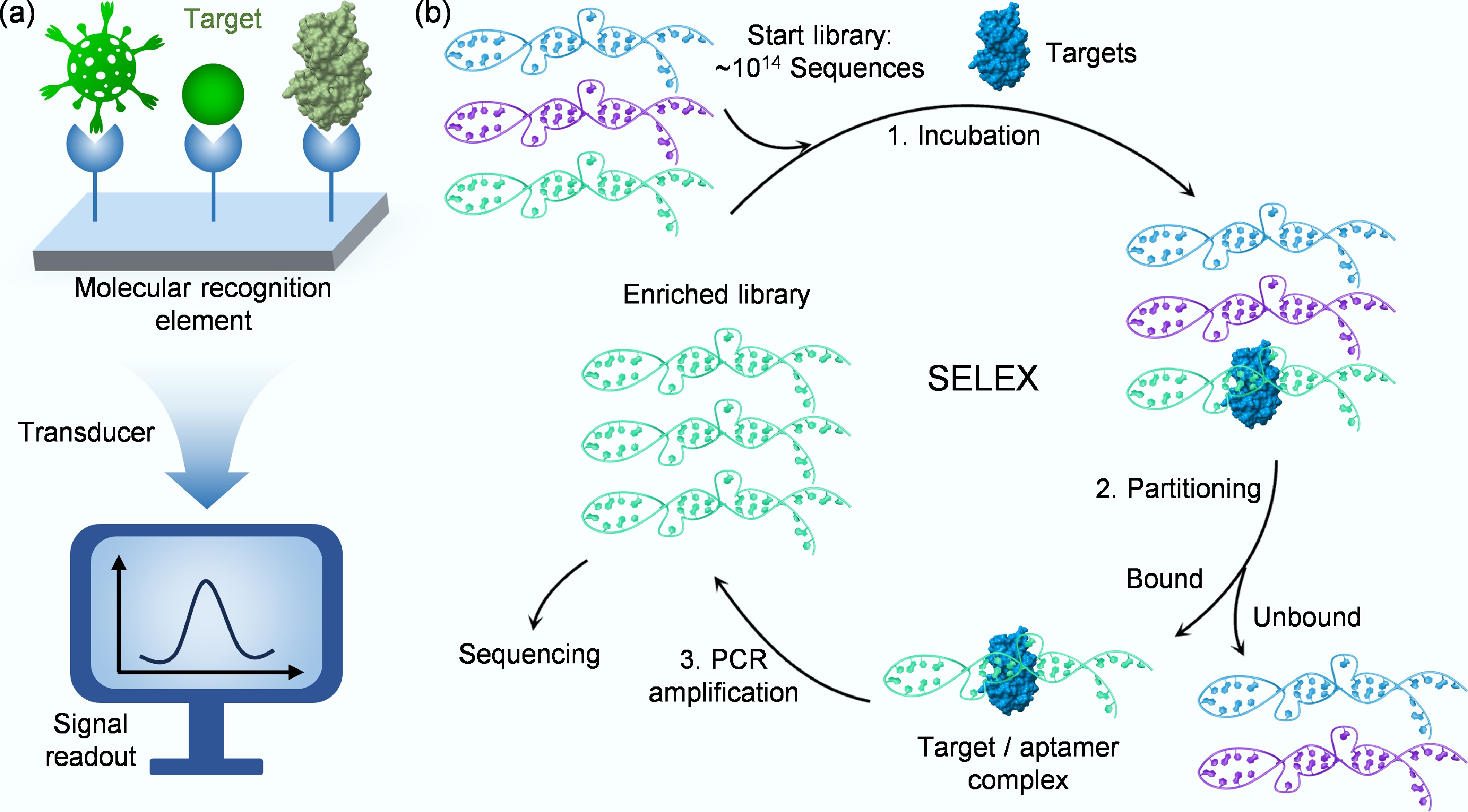
Figure 1.
(a) Schematic of a biosensor's key components for detecting target analytes. (b) Schematic illustration of the key steps in SELEX.
-
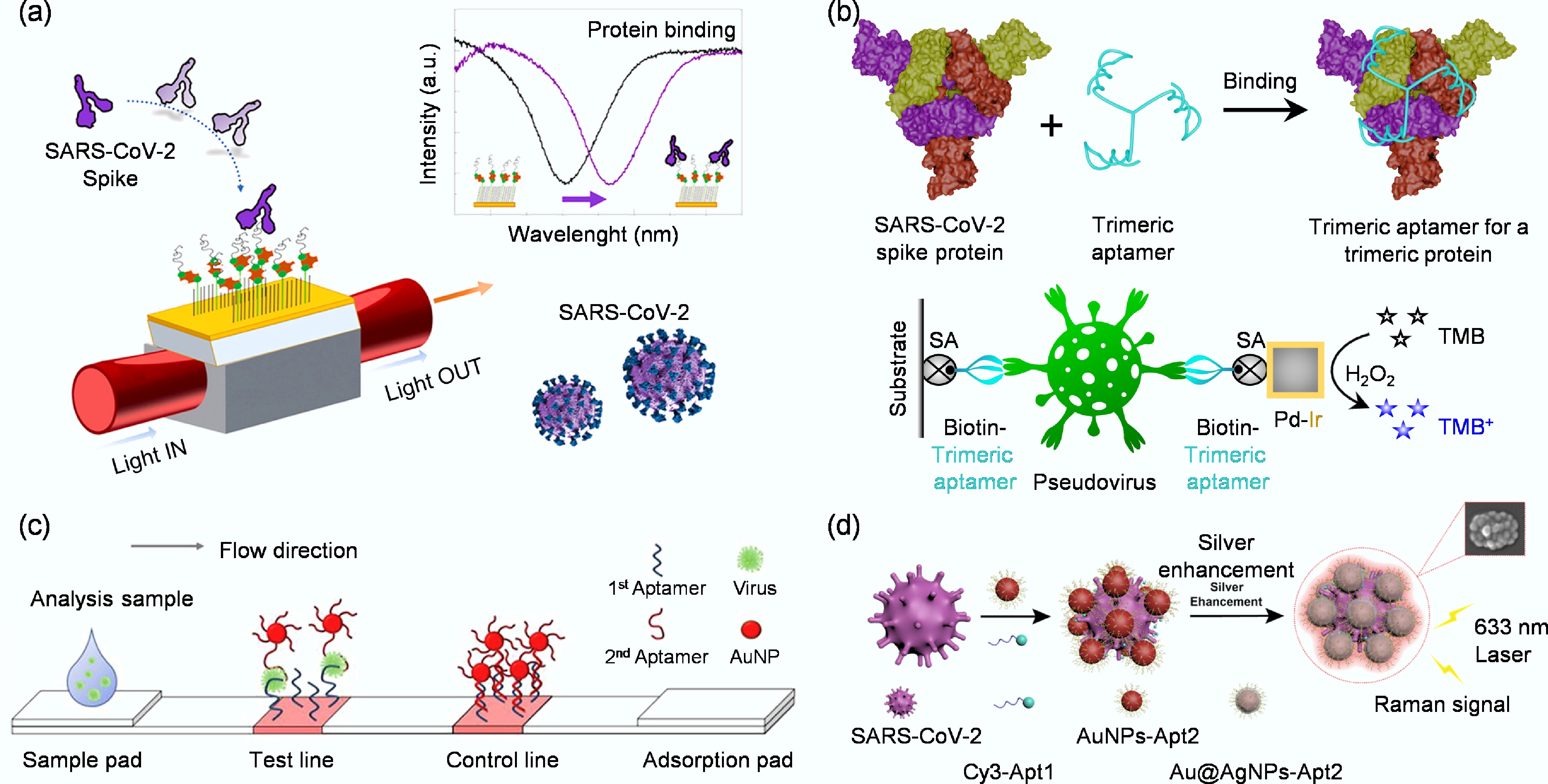
Figure 2.
(a) Schematic representation of a highly specific and sensitive SARS-CoV-2 spike protein biosensor[33]. (b) Design of an enzyme-linked aptamer binding assay for colorimetric detection of SARS-CoV-2 using trimeric aptamer TMSA52[34]. (c) Diagram of the whole virus particle detection system using a cognate pair of aptamer-based lateral flow strip[35]. (d) Schematic diagram of SERS aptamer biosensor for SARS-CoV-2 detection by assembling hotspots via S proteins on the surface of SARS-CoV-2[36].
-
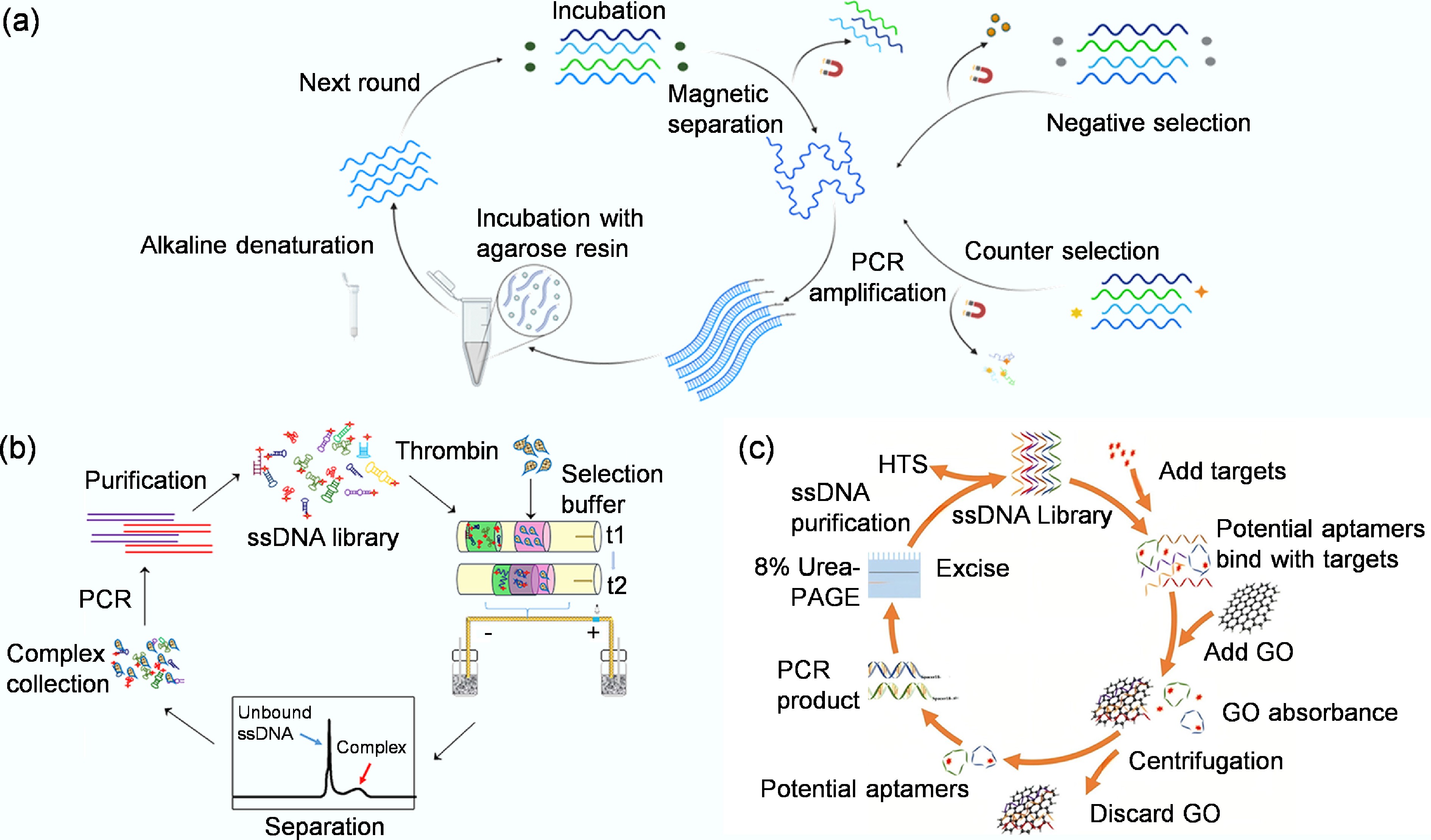
-
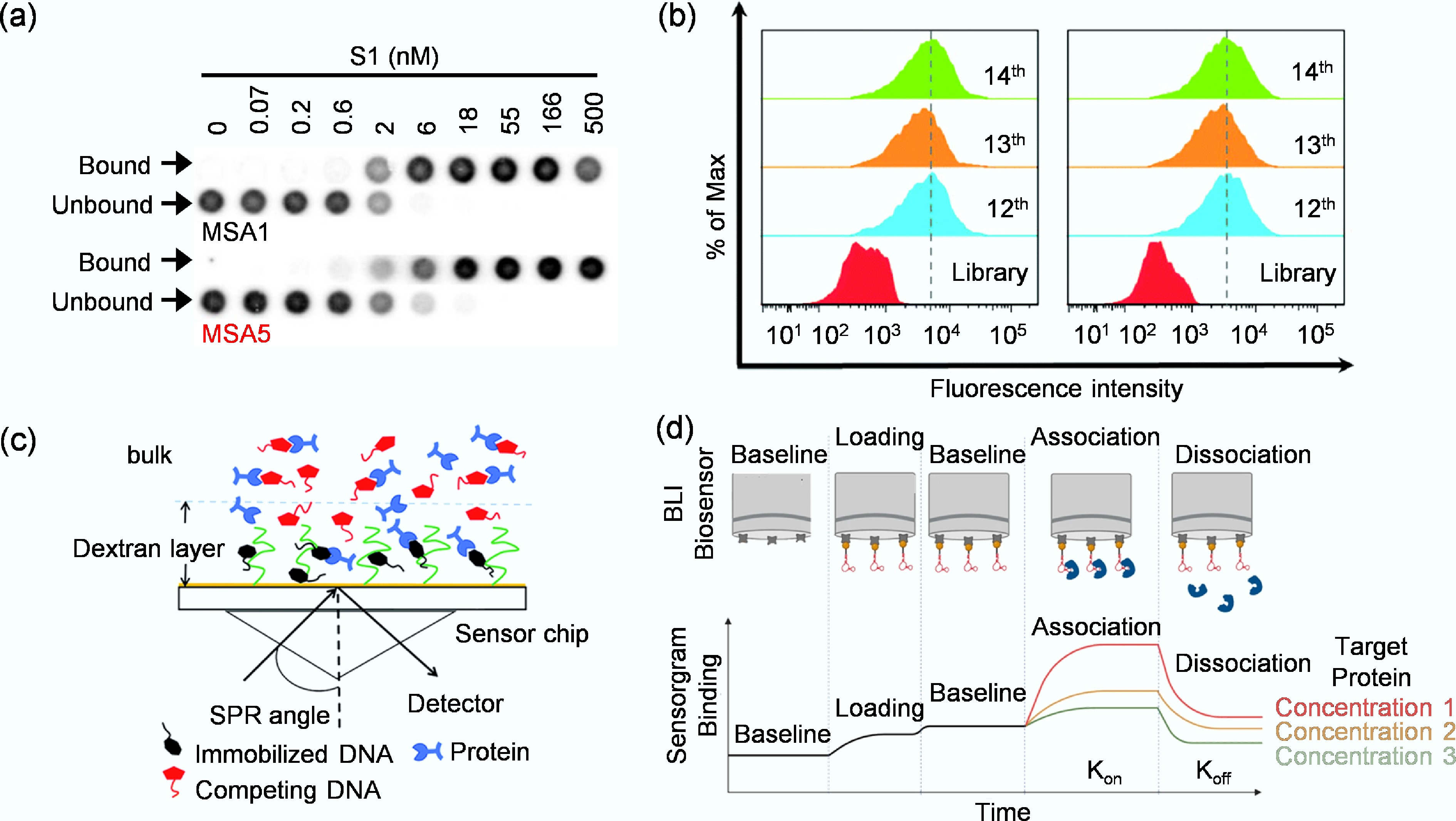
Figure 4.
(a) Assessment of binding affinity of MSA1 and MSA5 to the SARS-CoV-2 S1 protein by dot blot assay[57]. (b) Monitoring the binding ability of the enriched libraries to target KYSE150 cells and control KYSE30 cells by flow cytometry[59]. (c) Working principle of SPR for target binding analysis[60]. (d) The binding of a target protein to an immobilized aptamer ligand induces a change in the interference pattern[61].
-
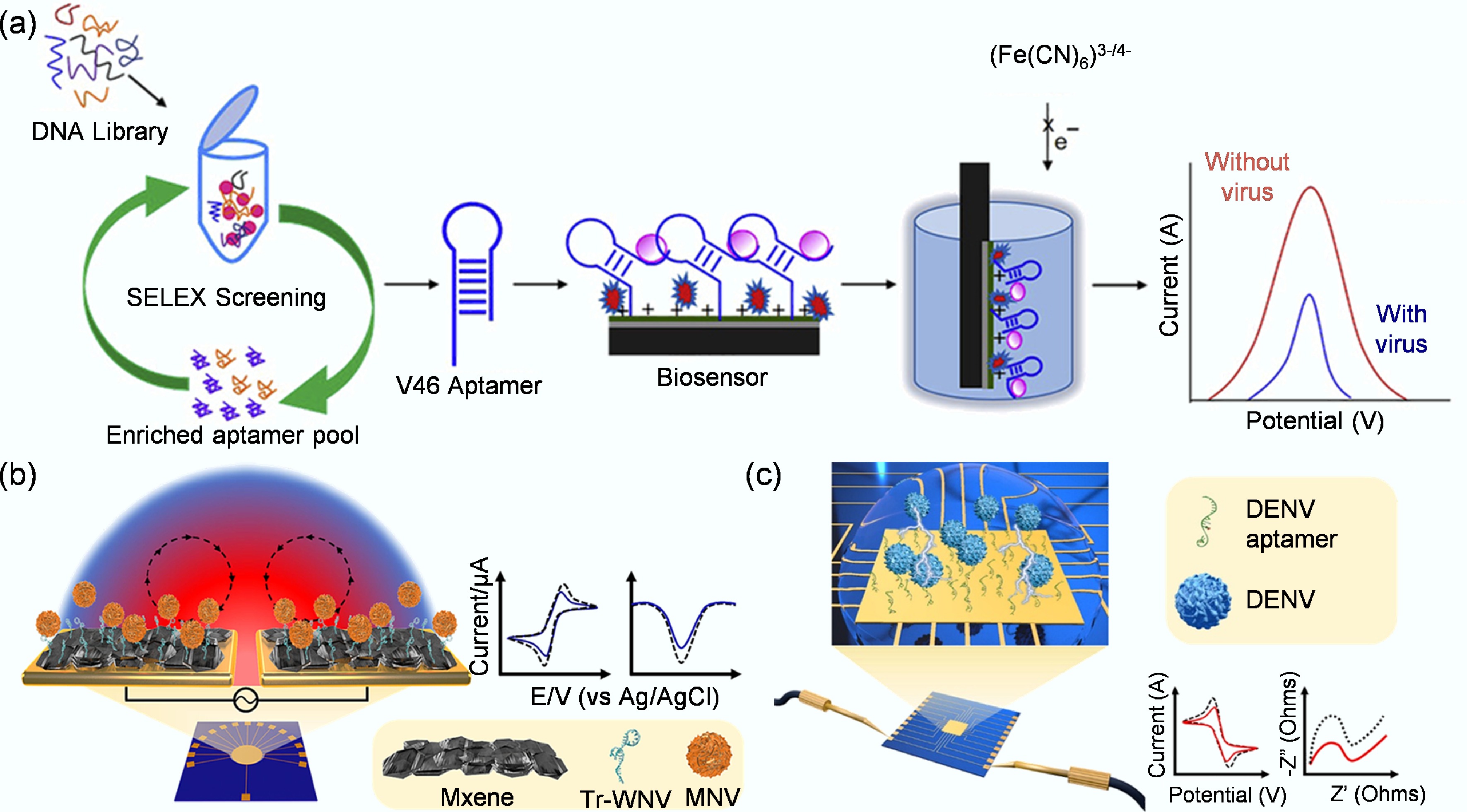
Figure 5.
(a) Label-free electrochemical aptamer biosensor based on differential pulsed voltammetry for the detection of H1N1 virus[99]. (b) Schematic representation of a rapid electrochemical biosensor for West Nile virus detection, featuring an MXene/Tr-WNV aptamer complex and fabricated by ACEF technology[75]. (c) Schematic representation of a biosensor for rapid electrochemical dengue virus detection by ACEF technology[76].
-
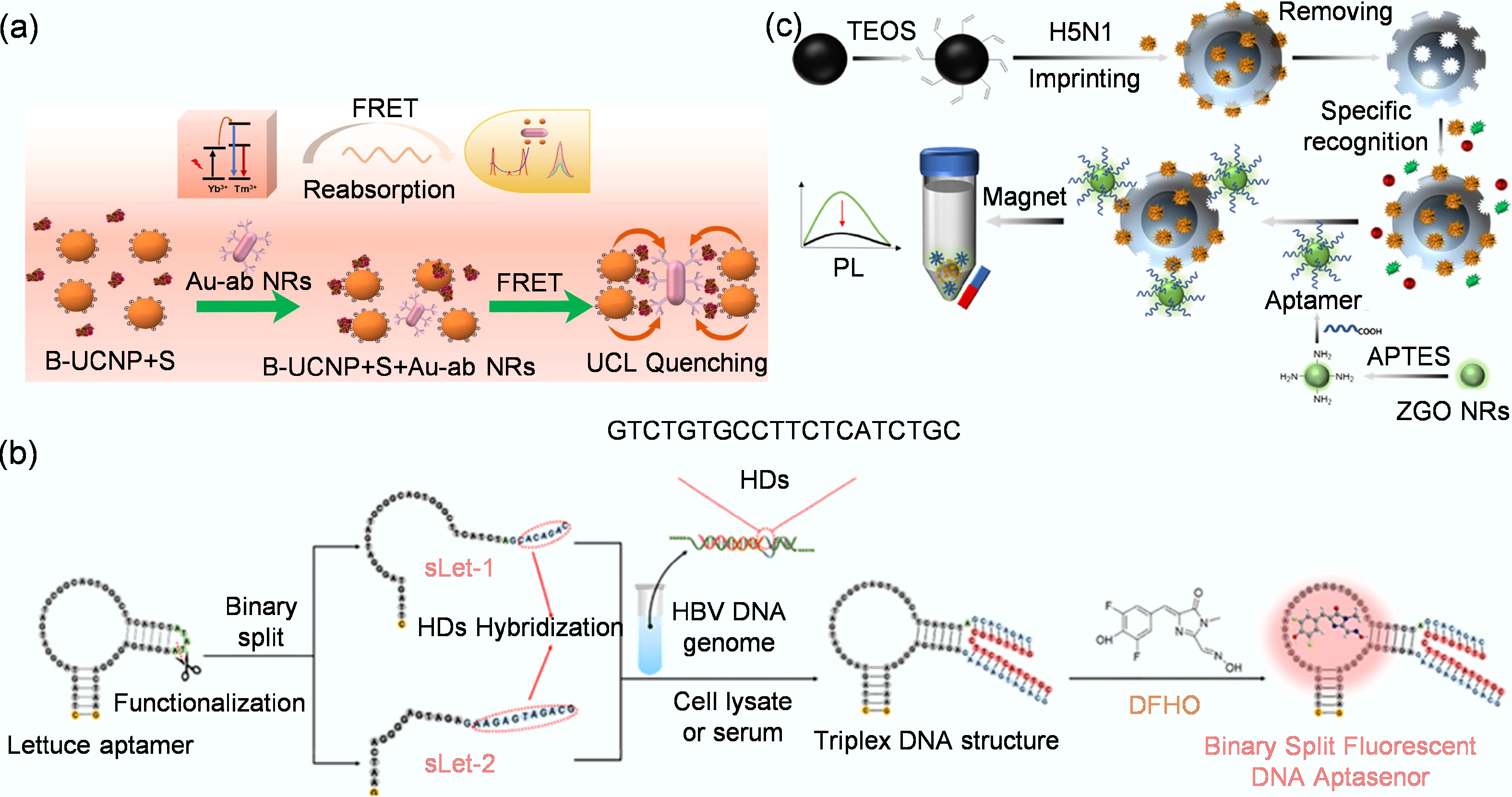
Figure 6.
(a) Detection system that utilizes the FRET effect to achieve highly sensitive, rapid, quantitative, and on-site detection of the spike protein of SARS-CoV-2[114]. (b) Schematic diagram for detecting HBV DNA fragments using the bifurcated fluorescent DNA aptamer biosensor[82]. (c) The preparation of MIP-aptamer biosensor for H5N1 detection[113].
-
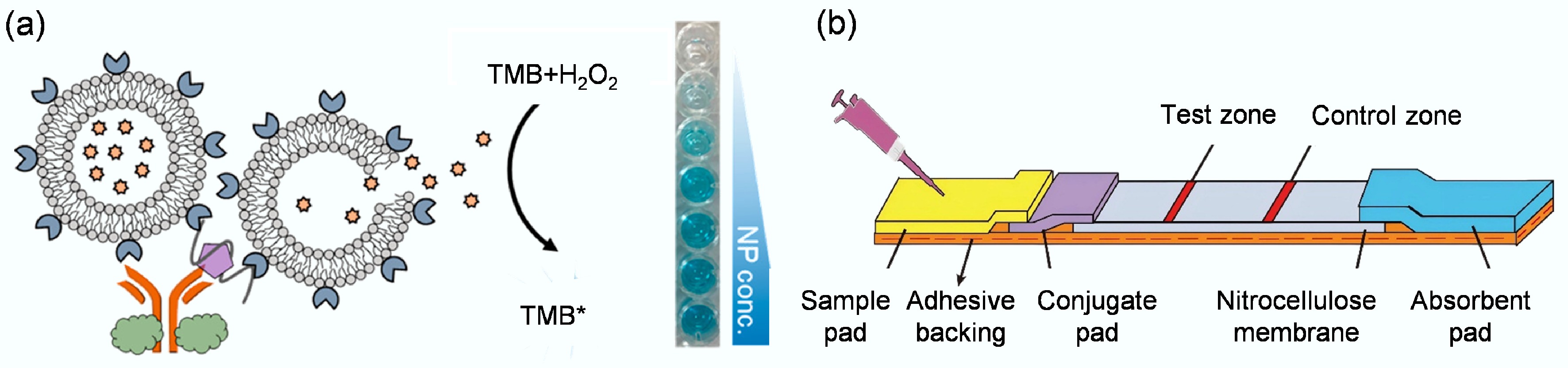
Figure 7.
(a) Depiction of a colorimetric biosensor designed to target the nucleocapsid protein of the severe fever with thrombocytopenia syndrome virus[88]. (b) Aptamer-based LFD that integrates strand-displacement amplification with a gold nanoparticle probe for highly sensitive detection of Siniperca chuatsi iridovirus[120].
-
-
Virus Target Detection method Aptamer name Kd LDR LOD Detection time HIV HIV-p24 Electrochemical[67] − − 0.93–93 μg/mL 51.7 pg/mL 30–60 min Zika Zika SF9 envelope protein Electrochemical[68] Zika–aptamer − − 2.4 × 107 Viruses 7.45 min YFV NS1 protein Electrochemical[69] YFV-37 aptamer 119.88 ± 27.93 nM − 2.366 pM 3 h HAdV HAdV-2 (VR-846) Electrochemical[70] HAdV-Seq4 0.9 ± 0.1 nM − 1 pfu/mL 30 min HNV (GII. 2) NoV VLP Electrochemical[71] APTL-1 148.13 ± 6.53 nM 1 ng/mL–
10 μg/mL0.28 ng/mL 30 min MNV (GII. 3) Capsid protein Electrochemical[72] AG3 − 20–120 aM 10 aM or 180 virus particles 60 min HPV-16 HPV-16 L1 Electrochemical[73] HPV-16 L1 aptamer − 0.2–2 ng/mL 0.1 ng/mL or
1.75 pM− HPV-16 HPV-16 E7 proteins Electrochemical[74] Sc5-c3 − − 100 pg/mL or
1.75 nM− WNV WNV envelope proteins Electrochemical (ACEF)[75] Tr-WNV aptamers 63.94 ± 11.05 nM − 1.06 pM 10 min DENV Virus envelope protein Electrochemical
(ACEF)[76]Isol-7 aptamer 261.1 ± 12.9 nM − 76.7 fM 10 min SARS-CoV-2 Pseudotyped SARS- CoV-2 virus Electrochemical[70] SARS2-AR10 − − 1 × 104 copies/mL 30 min SARS-CoV-2 Spike protein Electrochemical
(EIS)[77]DSA1N5 0.12 ± 0.02 nM − 1,000 VP/mL 5 min SARS-CoV-2 Spike protein Electrochemical[78] CoV2-RBD-1C 5.8 nM 0.5–8 μg/mL 72 ng/mL 70 min SARS-CoV-2 Nucleocapsid protein Fluorescence[79] NP-A48, NP-A58 − 10 fg–1.0 ng 5.87 pg 30 min H5N1 Magnetic MIPs Fluorescence[10] ZGO-H5N1 Apt − − 0.0128 HAU/mL 15 min SMV (GII. 2) Capsid proteins Fluorescence[80] Aptamers 25 232 nM 1–5 µg/mL 1 log10 RNA copies per 3 g lettuce 15 min HBV e antigen Fluorescence[81] − − 42–420 nM 26.5 nM
(609 ng/mL)2 min HBV HDs Fluorescence[82] sLet-1, sLet-2 − − 0.0009 μM 4 min HPV-16 HPV-16 L1 Fluorescence[83] S4 10−99 nM 10−1−104 ng/mL 0.1 ng/mL > 240 min SARS-CoV-2 Nucleocapsid protein Fluorescence
(PIFE)[84]PCL-Apto3 − 1–40 nM 0.05 ng/μL 1–2 min SARS-CoV-2 Spike protein Fluorescence
(NSET)[85]CoV2-RBD-4C 19.9 nM 10–500 virus/mL 130 fg/mL (antigen), 8 particles/mL (virus) 10 min HBV Surface antigen Fluorescence[49] H01, H02, H03 − 1–200 ng/mL 0.1 ng/mL − ASFV p30 Colorimetric[86] Atc-20 140 ± 10 pM − 0.61 ng/mL − BVDV Glycoprotein E2 Colorimetric[87] Apt 31 − − 0.27 copies/mL 15–20 min SFTS virus Nucleocapsid protein Colorimetric[88] SFTS-apt3 0.8 ± 0.2 μM − 0.009 ng/mL 60 min SARS-CoV-2 Spike protein Colorimetric[89] MBA5SA1 0.12 nM − − 30 min SARS-CoV-2 Spike protein Colorimetric[90] TMSA52 8.8–23.7 pM − 3.2 × 103 cp/mL 60 min SARS-CoV-2 Spike protein Colorimetric[91] TMSA5 6.8–14.8 pM − 4 ×105 cp/mL 30 min H5N2 Virus particles Colorimetric[35] JAPT, JHAPT − − 2.09 × 105 EID50/mL 20 min HIV HIV-1 gp120 Colorimetric[92] HD2, HD3, HD4, HD5 − 1–1,000 ng/mL 8.0 ± 1.2 ng/mL 5–10 min HIV HIV-1 gp120 Colorimetric[93] B40t77 RNA aptamer − 0.1–8 μg/mL 0.2 μg/mL 60 min HIV HIV-I Tat Colorimetric[94] − 120 pM 1.0–500 nM 1 pM (1.5 pg/mL) − Influenza B Nucleoprotein Colorimetric[95] INFA-apt4 77.6–125.7 nM 1–105 pg/mL 0.16 pg/mL − ASPV Coat proteins (PSA-H, MT32) SPR[96] − 55, 8 nM − − − SARS-CoV-2 Spike protein SPR[66] CoV2-RBD-1C 5.8 nM − 36.7 nM 10 min H5N1 HA SPR[97] − 4.65 nM 0.128–1.28 HAU − 1.5 h Noroviruses
(GII. 4)VP1 SPR[98] Aptamer I−IV − 70–200 aM 70 aM > 60 min SARS-CoV-2 Spike protein SERS[36] Apt1-Cy3
Au@AgNPs-Apt2− − 5.26 TCID50/mL 10 min H1N1 HA SERS[99] V46, V57 19.2, 29.6 nM 1–106 PFU/mL 0.62 HAU/mL − H1N1 A/H1N1 target SERS[100] Cy3-labeled aptamers − − 97 PFU/mL 20 min H1N1 HA SERS[101] RHA0385 2–5 nM − 2 × 105 VP/mL 15 min LDR: Linear Detection Range; LOD: Limit Of Detection; ASFV: African Swine Fever Virus; BVDV: Bovine Viral Diarrhea Virus; SFTS: Severe Fever With Thrombocytopenia Syndrome; H5N1: Avian Influenza Virus H5N1; HA: Hemagglutinin; HAU: Hemagglutination Units; ASPV: Apple Stem Pitting Virus; BPMV: Bean Pod Mottle Virus; HIV: Human Immunodeficiency Virus; Tat: Trans-Activator of Transcription; WNV: West Nile Virus; ACEF: Alternating Current Electrothermal Flow; NSET: Nanoparticle Surface Energy Transfer; PIFE: Protein-Induced Fluorescence Enhancement; EIS: Electrochemical Impedance Spectroscopy; FRET: Fluorescence Resonance Energy Transfer; DENV: Dengue Virus; YFV: Yellow Fever Virus; PBS: Phosphate-Buffered Saline; HAdV: Human Adenovirus; H5N2: Avian Influenza Virus H5N2; LFD: Lateral Flow Devices; H1N1: Influenza A H1N1 virus; SERS: Surface-Enhanced Raman Scattering; HBV: Hepatitis B Viruses; HDs: HBV DNA segment complementary sequences; HNV: Human Norovirus; MNV: Murine Norovirus; SMV: Snow Mountain Virus; '−' indicates data not available. Table 1.
Summary of aptamer affinity and biosensor performance for viral detection
-
Biosensor type Typical sensitivity Detection time Cost Portability Multiplexing Key advantages Major limitations Electrochemical fM–pM Minutes Low High Moderate High sensitivity, portability,
low cost, miniaturizableSusceptible to electrode fouling in complex samples Fluorescent pM–nM Minutes to hours Low-moderate Moderate High High sensitivity, versatile designs, suitable for imaging Background fluorescence requires a light source/detector Colorimetric pM–nM Minutes Very low Very high Low Naked-eye readout, simple,
low cost, ideal for POCTLower sensitivity, semi-quantitative SPR pM–nM Real-time High Low Moderate Label-free, real-time kinetics, high-information content Bulky equipment, high cost, sensitive to bulk refractive index changes SERS fM - single virus Minutes Moderate-high Moderate High Ultra-high sensitivity, fingerprinting, multiplexing Signal heterogeneity, substrate reproducibility Table 2.
Comparative analysis of aptamer-based biosensors for viral detection
Figures
(8)
Tables
(2)