-
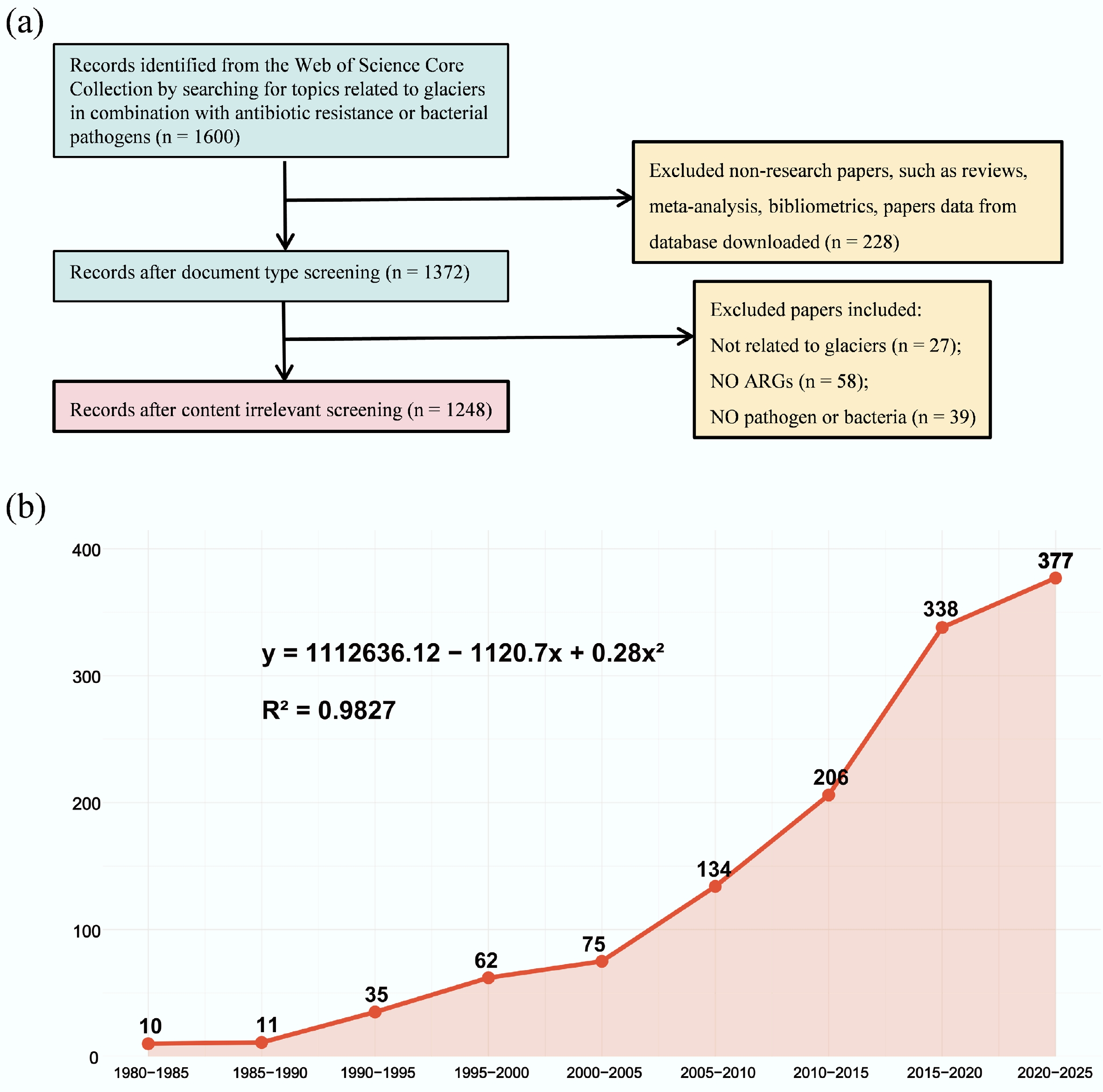
Figure 1.
(a) Number of documents at each stage of the screening process. (b) Development trend of papers and research fields from 1980 to 2025.
-
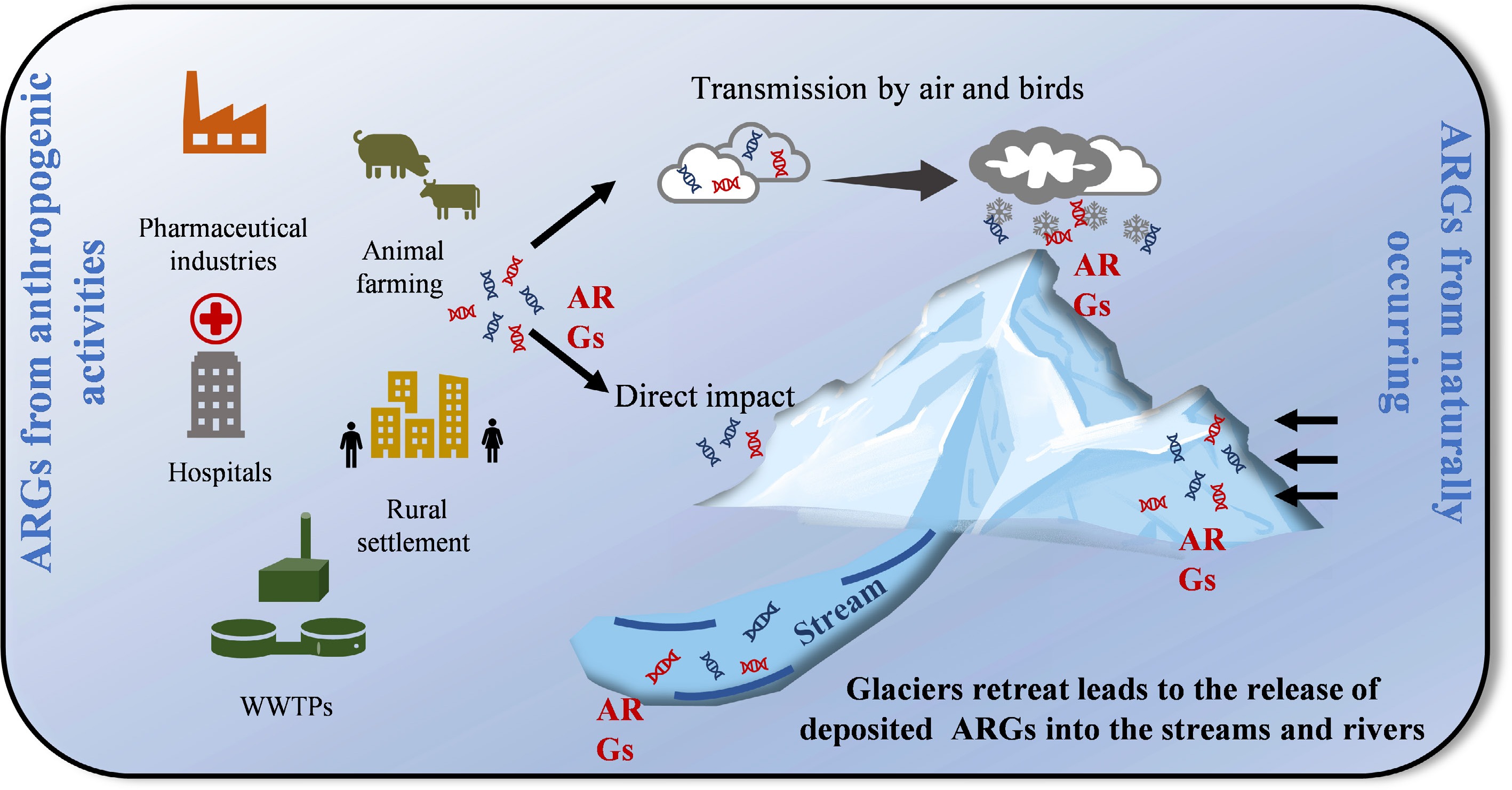
Figure 2.
Sources of ARGs in glaciers: natural legacy and anthropogenic inputs.
-

Figure 3.
The glacier continuum is a key pathway for downstream transmission of ARGs.
-
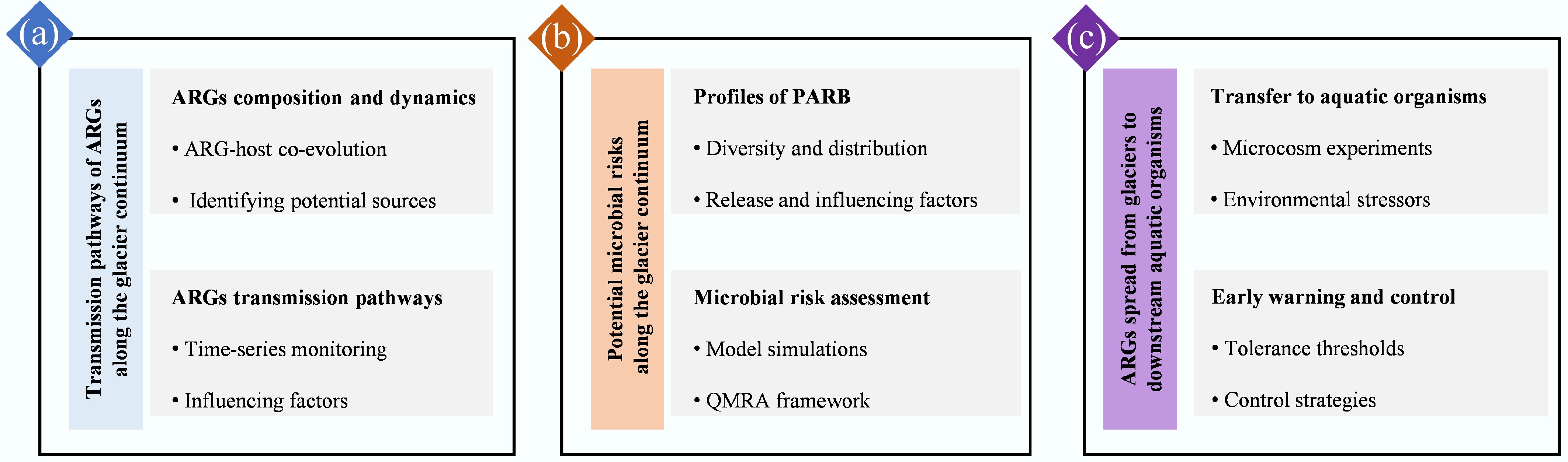
Figure 4.
Schematic overview of the technical route for future study.
-
Class Mechanism type Gene type Location Approach Ref. Aminocoumarin Drug efflux mdtB King George Island, West Antarctica Metagenomic analysis [36] mdtC Abisko, Sweden Metagenomic analysis [37] Aminoglycosides Drug inactivation aac, ant Mackay Glacier, South Victoria Land, Antarctica Metagenomic analysis [20] aac, aph Chongce glacier, and Eboling mountain, Xizang Metagenomic analysis [38] aac(6')-Ib3, aph(2')-Id Tianshan Mountain Urumqi No.1 glacier Metagenomic analysis [39] Drug efflux amrB King George Island, West Antarctica Metagenomic analysis [36] Beta-lactam Drug efflux acrB, mexB, tolC Lake Namco, Qiangyong glacier, Xizang Metagenomic analysis [33] Drug inactivation Class A: bla(GES, VEB, NPS, CTX, PSE);
Class C: bla(EC);
Class D: bla(OXA)Chongce glacier, and Eboling mountain, Xizang Metagenomic analysis [38] Target replacement mecA River Lena region in Central Yakutia, Russia Bacterial genomic sequencing [40] Bacitracin Target modification bacA, bcrA Chongce glacier, and Eboling mountain, Xizang Metagenomic analysis [38] bacA King George Island, West Antarctica Metagenomic analysis [36] Glycopeptide Target modification vanHA Bear Creek, Yukon, Canada Metagenomic analysis [17] vanR McMurdo Dry Valleys, East Antarctica Microarray GeoChip 4.0 [41] Macrolides Drug efflux macB King George Island, West Antarctica Metagenomic analysis [36] oleC Kongsfjorden Region of Svalbard in the High Arctic SmartChip Real-Time PCR [42] Target protection ermC River Lena region in Central Yakutia, Russia Bacterial genomic sequencing [40] ermC Northern Bering Sea qPCR [43] Drug inactivation mph River Lena region in Central Yakutia, Russia Bacterial genomic sequencing [40] MLSB (macrolide-lincosamide-streptogramin B) Target protection msrA River Lena region in Central Yakutia, Russia Bacterial genomic sequencing [40] Target modification erm(K, 34, 36) Kongsfjorden Region of Svalbard in the High Arctic SmartChip Real-Time PCR [42] Quinolones Drug efflux qepA Northern Bering Sea qPCR [43] Target protection qnr(A, B, D, S) Northern Bering Sea qPCR [43] Target modification gyrA Abisko, Sweden Metagenomic analysis [37] Sulfonamides Target replacement sul(1, 2, 3) Northern Bering Sea qPCR [43] sul(1, 2) Chongce glacier, and Eboling mountain, Xizang Metagenomic analysis [38] Target modification folP Abisko, Sweden Metagenomic analysis [37] Tetracyclines Drug efflux tet(A, B, C, D) Northern Bering Sea qPCR [43] tet(D, L, G, Y, 33, 39) Chongce glacier, and Eboling mountain, Xizang Metagenomic analysis [38] Target protection tet(M) Northern Bering Sea qPCR [43] tet(M, O, 36) Chongce glacier, and Eboling mountain, Xizang Metagenomic analysis [38] Trimethoprim Target replacement dfrA Mackay Glacier, South Victoria Land, Antarctica Metagenomic analysis [20] dfrE King George Island, West Antarctica Metagenomic analysis [36] Multidrug Drug efflux acrE King George Island, West Antarctica Metagenomic analysis [36] acr(A,R) Kongsfjorden Region of Svalbard in the High Arctic SmartChip Real-Time PCR [42] mepA Kongsfjorden Region of Svalbard in the High Arctic SmartChip Real-Time PCR [42] Table 1.
Examples of ARGs, resistance mechanisms, and drug classes detected in glacial environments worldwide
-
Method category Specific technique Principle Advantages Limitations Recommendations for standardization Culture-dependent Kirby-Bauer Disk Diffusion[81] Measures the zone of inhibition around the antibiotic disc Simple, low-cost, provides phenotypic resistance data Misses > 99% of unculturable microbes; low throughput Use as a complementary method to characterize isolated strains. Standardize media and incubation conditions Targeted molecular Quantitative PCR (qPCR)[82] Amplifies and quantifies specific, known ARG sequences using primers Highly sensitive and quantitative for targeted genes Limited to pre-selected targets; primer bias Use universal 16S rRNA gene normalization for bacterial abundance; Report detection limits; Use standardized primer sets when available High-Throughput qPCR (SmartChip)[83] Allows simultaneous quantification of hundreds of pre-defined ARGs High-throughput for targeted genes; broad profiling Still limited to pre-designed assays; high cost Normalize data per 16S rRNA gene. Clearly report the targeted gene panel Untargeted molecular Shotgun Metagenomics[84,85] Random sequencing of all DNA in a sample, followed by bioinformatic annotation Captures both known and novel ARGs; provides context (MGEs, hosts) Computationally intensive; depth of sequencing affects detection sensitivity Recommendation for primary use. Use deep sequencing. Normalize using reads per kilobase per million (RPKM/TPM) or cell count; Use standardized, curated databases (e.g., CARD, SARG); Deposit raw data in public repositories Emerging: Long-Read Sequencing (e.g., Nanopore, PacBio)[86] Sequences long DNA fragments Resolves complete ARG contexts (plasmids, chromosomes); links ARGs to hosts Higher error rate (Nanopore); lower throughput; higher cost Use for resolving genetic context and host association; Combine with short-read data for hybrid assembly for accuracy Table 2.
Comparison of methods for detecting ARGs in glacial environments
Figures
(4)
Tables
(2)