-
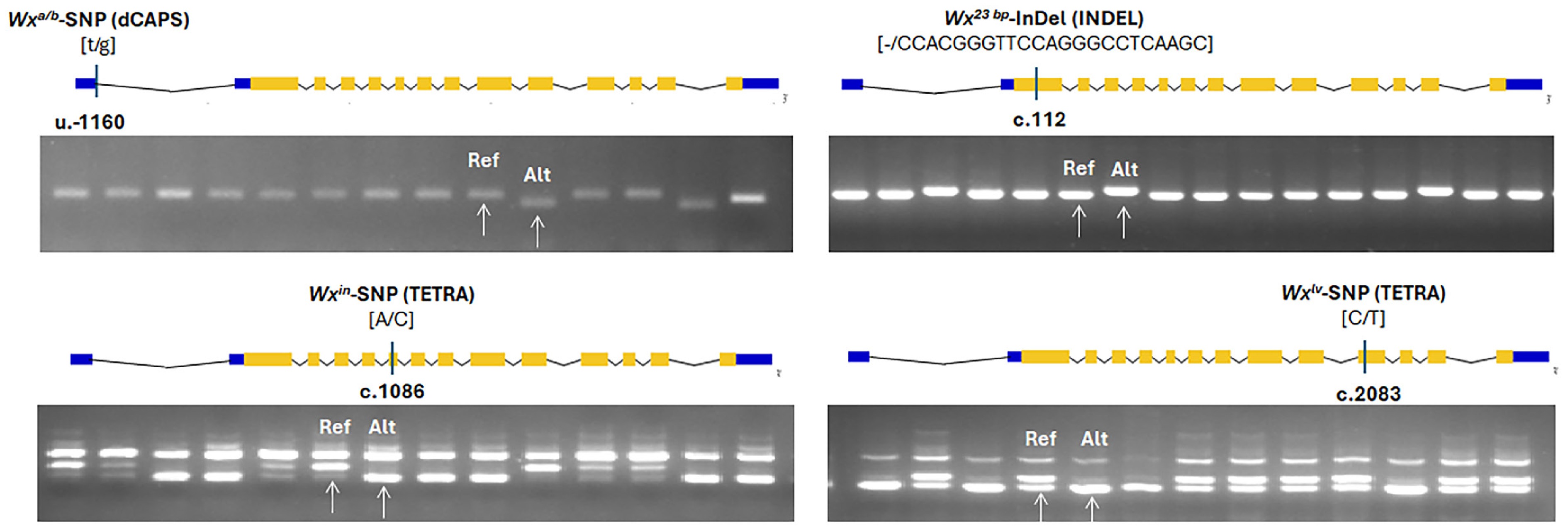
Figure 1.
Allele type and position of candidate FNPs of Waxy (Wxa/b, Wx23 bp, Wxin, Wxlv) alleles, and corresponding PCR results. The UTRs, exons, and introns of different genes are represented by blue boxes, orange boxes, and fold lines, respectively. The position of each variation is labeled as the relative distance to the ATG in the UTRs (u) and CDS (c). Allele variation of each gene is shown in the square bracket, with the front nucleotide variation representing Nipponbare (Reference) Allele and the back nucleotide variation representing non-NIP-type. The white arrows denote the band pattern of the reference (Ref) allele and alternative (Alt) allele, respectively.
-
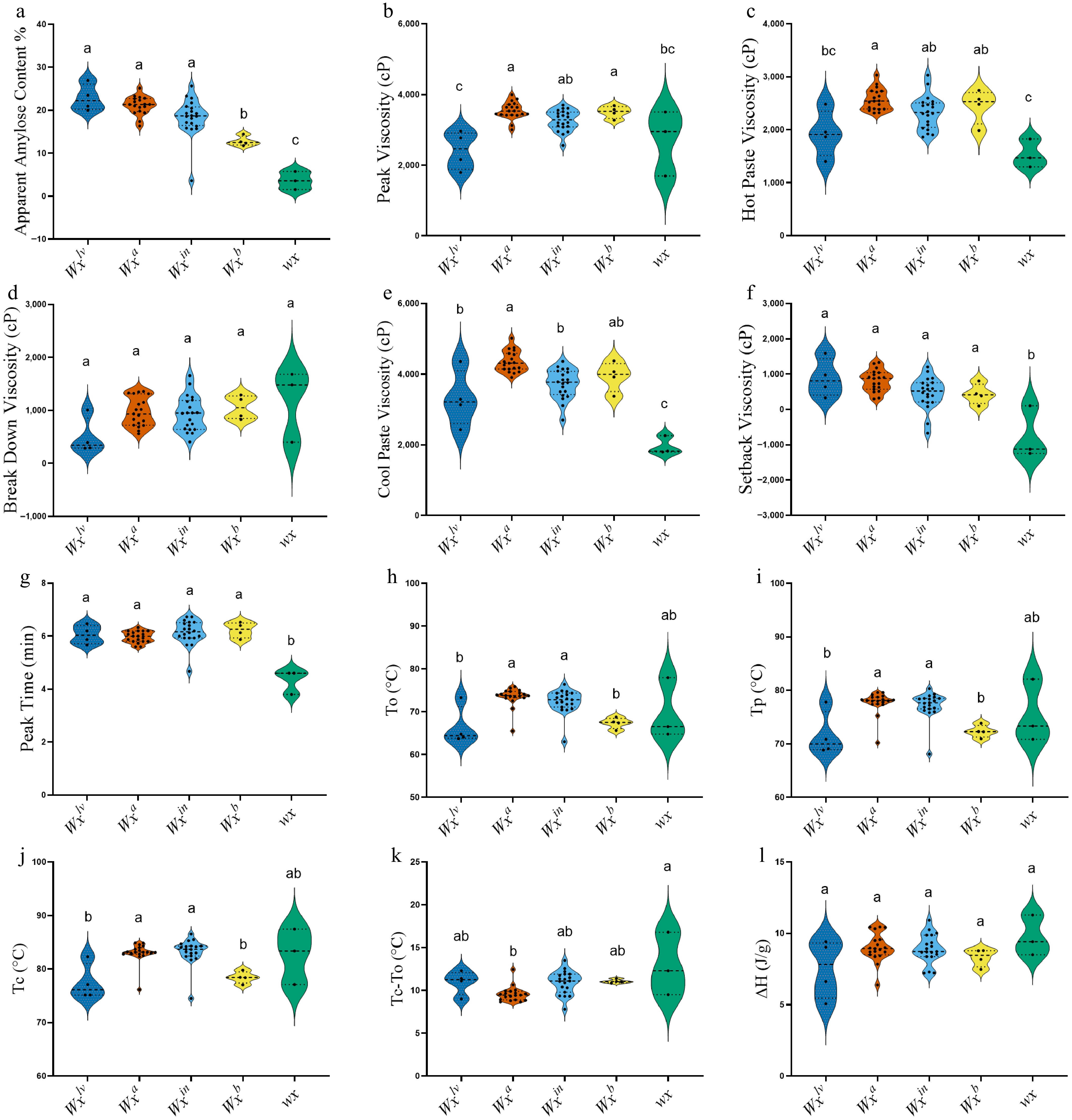
Figure 2.
Violin plot of Malaysian rice varieties grouped by different Wx alleles (Wxlv, Wxa, Wxin, Wxb, and wx), showing variation in (a) apparent amylose content, and (b) physicochemical parameters of RVA (Rapid Visco Analyzer), Peak Viscosity (PKV), (c) Hot Paste Viscosity (HPV), (d) Break Down Viscosity (BDV), (e) Cool Paste Viscosity (CPV), (f) Setback Viscosity (SBV), (g) Peak Time (PT), and DSC (differential scanning calorimetry), (h) To, (i) Tp, (j) Tc, (k) Tc–To, (l) ΔH (J/g). Significant statistical differences exist between groups with no shared character above the columns.
-
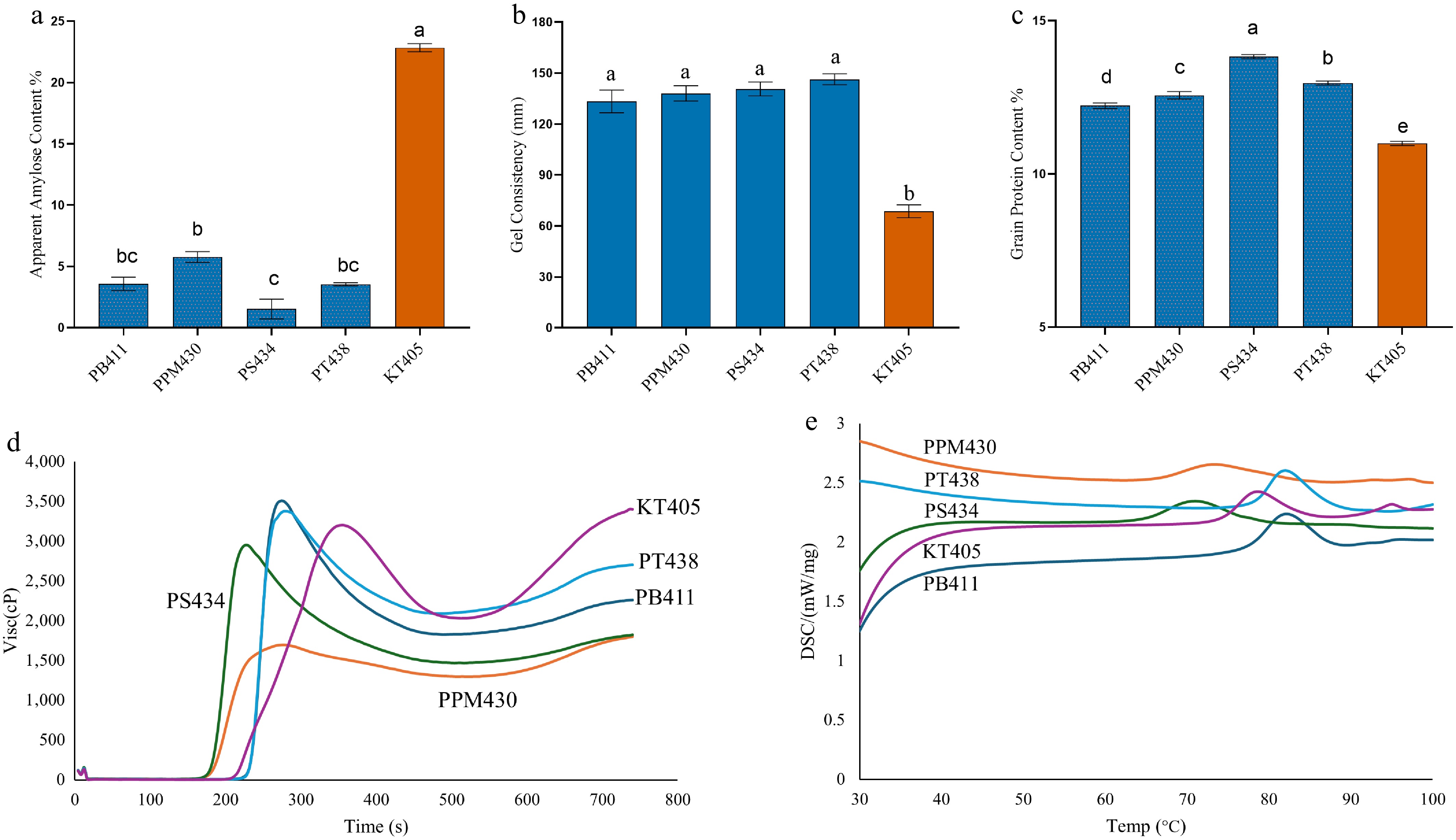
Figure 3.
(a) Apparent amylose content, (b) gel consistency, (c) grain protein content, (d) RVA (Rapid Visco Analyzer), and (e) DSC (Differential Scanning Calorimetry) graphs of Malaysian waxy rice varieties compared with KT405 (Wxin). PB411 (PULUT BURUNG), PPM430 (PADI PULOT MELAYANG), PS434 (PULUT SELEMYAM), PT438 (PULUTAN), KT405 (KATUNG). Significant statistical differences exist between groups with no sharing character above the columns (a and b).
-
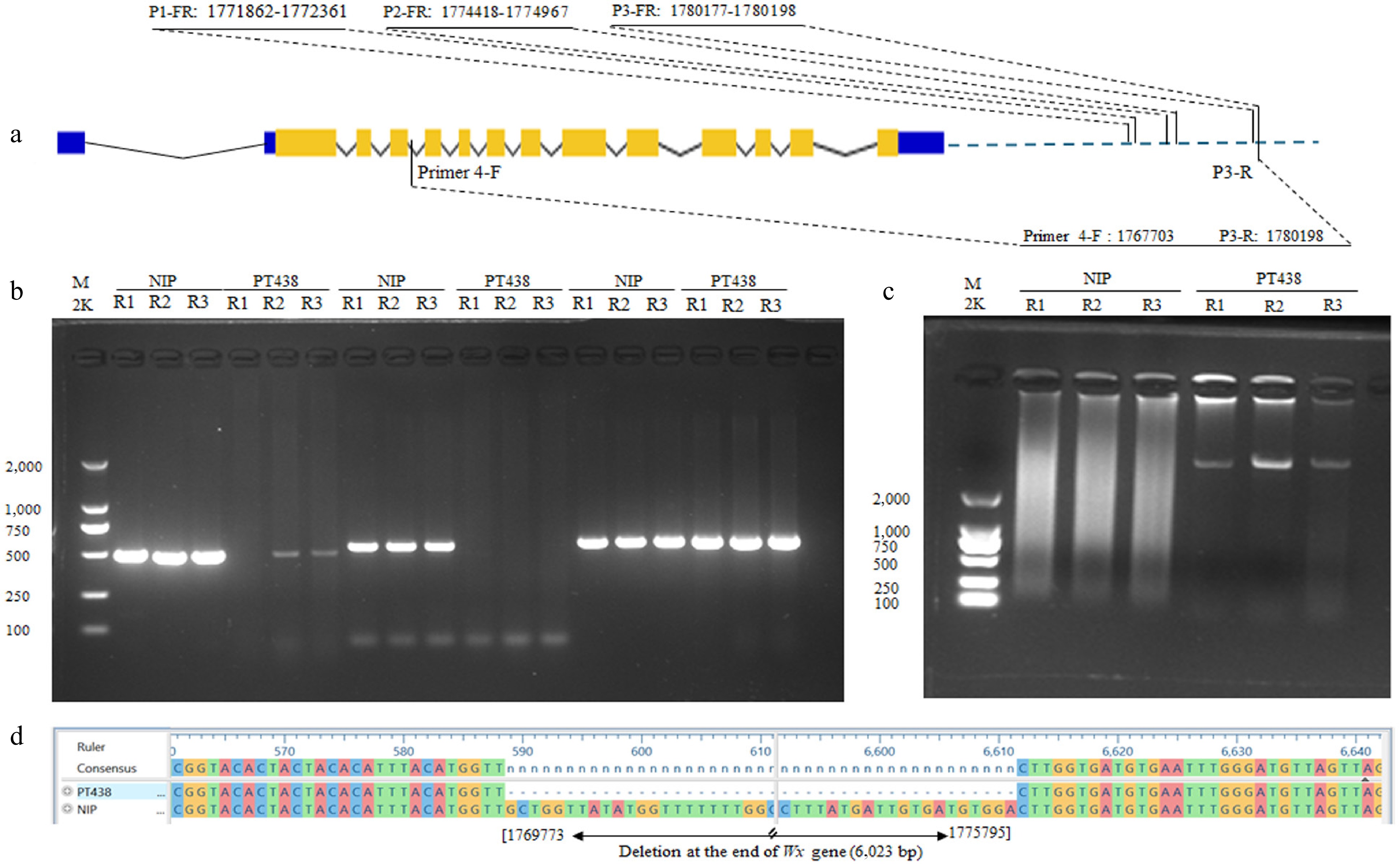
Figure 4.
(a) Deletion mapping, and (b) PCR amplification profile of the Malaysian rice variety PT438 (PULUTAN) using three primer pairs (P1-FR, P2-FR, and P3-FR) at the downstream of waxy gene. The position of each primer is shown on the gene structure, and NIP was amplified as a control. (c) and (d) are PCR amplification profiles and sequencing results showing a 6,023 bp deletion at positions 1769773−1775795 of the Malaysian rice variety PT438 (PULUTAN) compared with NIP. The promoters, UTRs, exons, and introns of the Waxy gene are represented by blue lines, blue boxes, orange boxes, and hat lines, respectively. R1, R2, and R3 are three repeats for each PCR amplification profile.
-
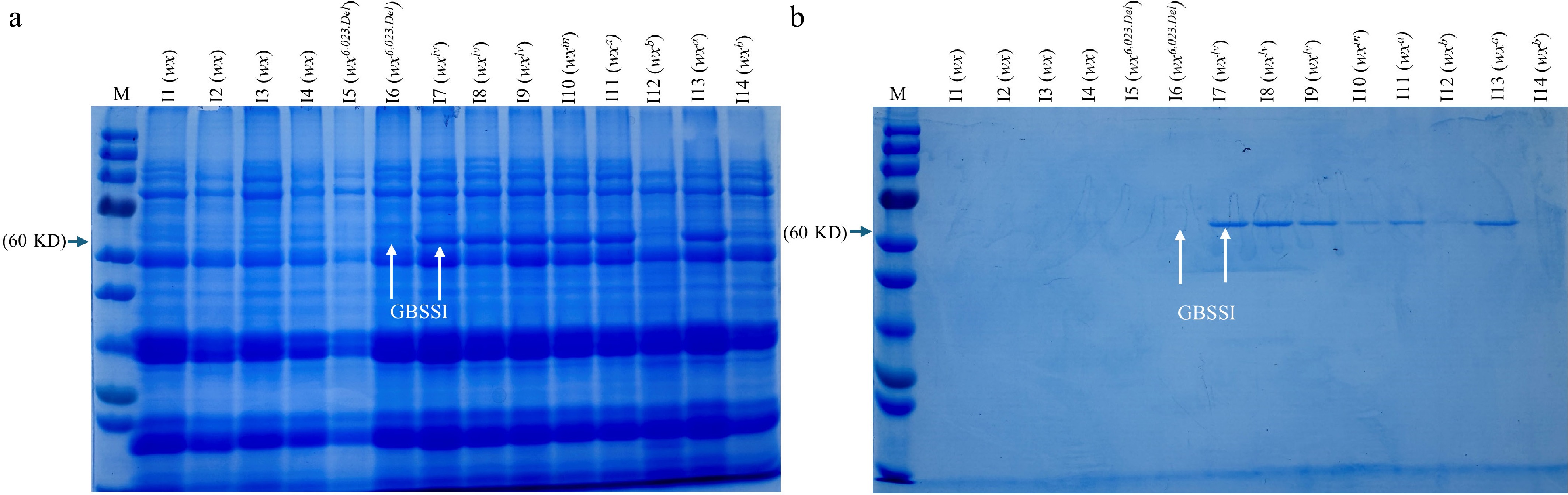
Figure 5.
SDS–PAGE assay of (a) total grain proteins and (b) starch granule-bound GBSSI in mature grains of NIP (I1 with Wxb), PS434 (I2 with wx), PB411 (I3 with wx), PPM430 (I4 with wx), PT438 (I5&I6 with wx6,023.Del), MR167 (I7 with Wxlv), MR 84 (I8 with Wxlv), PL425 (I9 with Wxlv), KT405 (I10 with Wxin), MR69 (I11 with Wxa), MR185 (I12 with Wxb), ZJ116 (I13 with Wxa), 9311 (I14 with Wxb). White arrows denote the protein level of wx6,023.Del and Wxlv genotype.
Figures
(5)
Tables
(0)