-
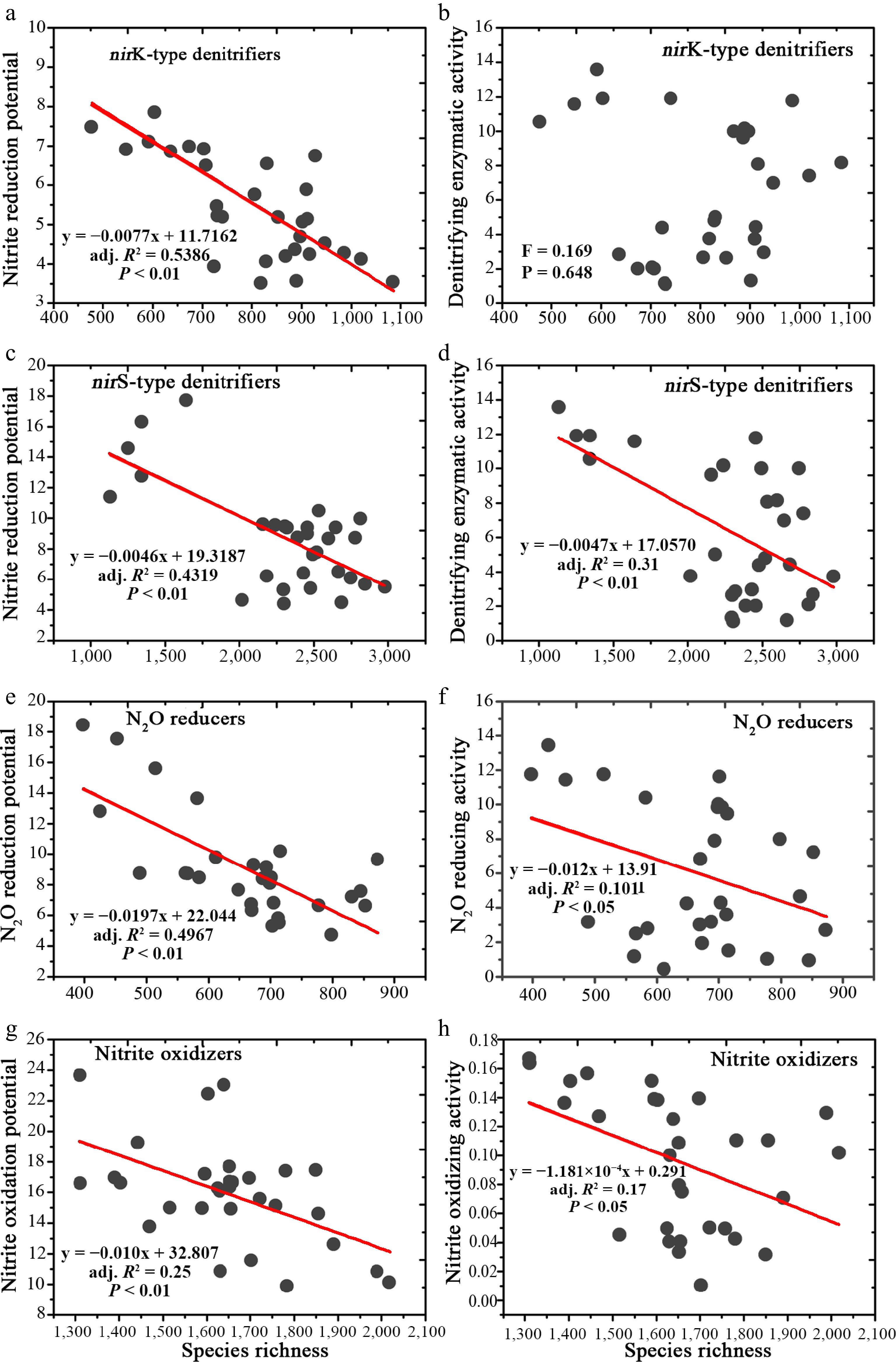
Figure 1.
Linear regressions showing the relationship between species richness and N-transforming capability.
-
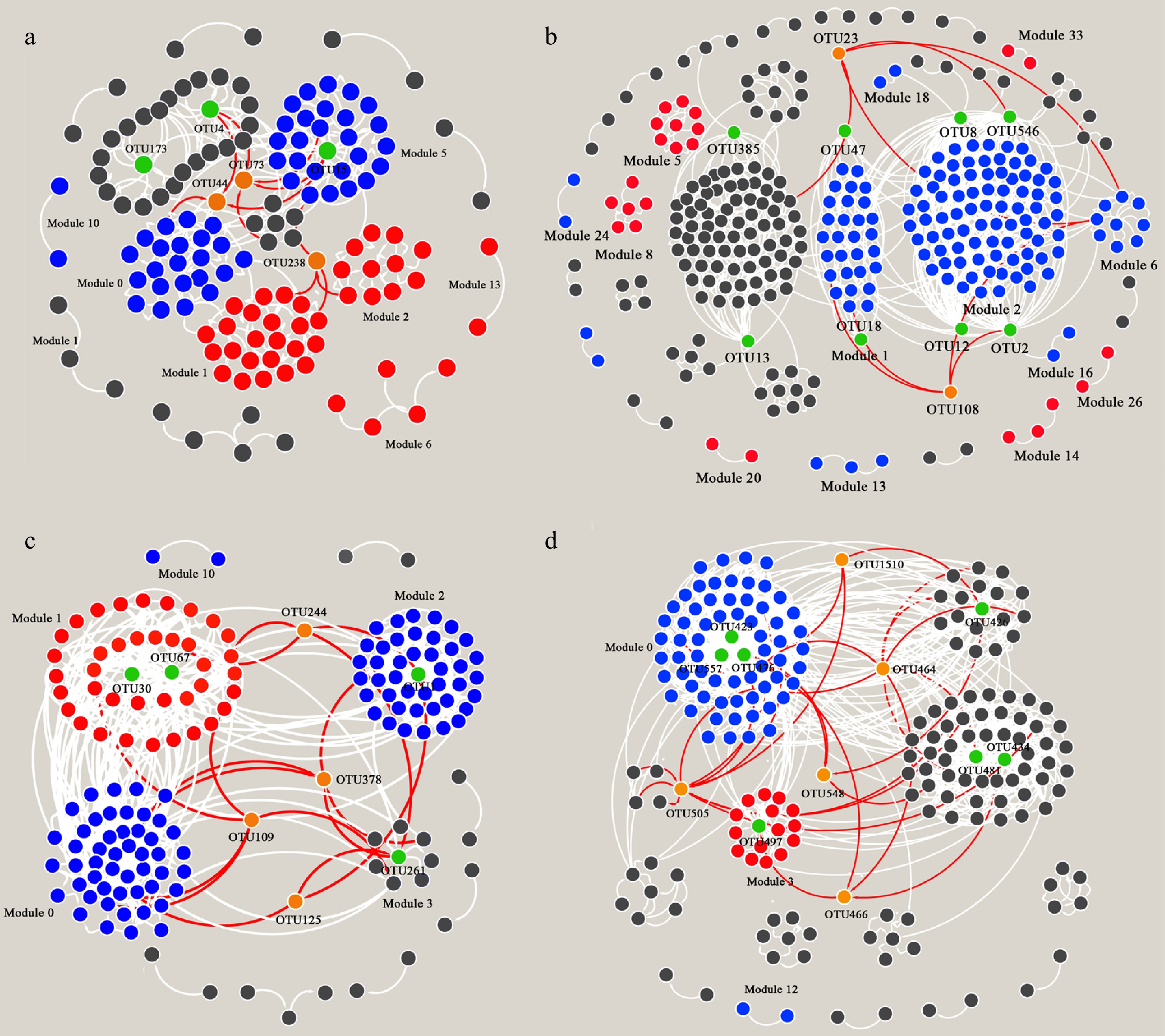
Figure 2.
Co-occurrence networks based on Pearson correlation for (a) nirK-type denitrifying bacteria, (b) nirS-type denitrifying bacteria, (c) N2O reducing bacteria, and (d) Oxidizing bacteria.
-
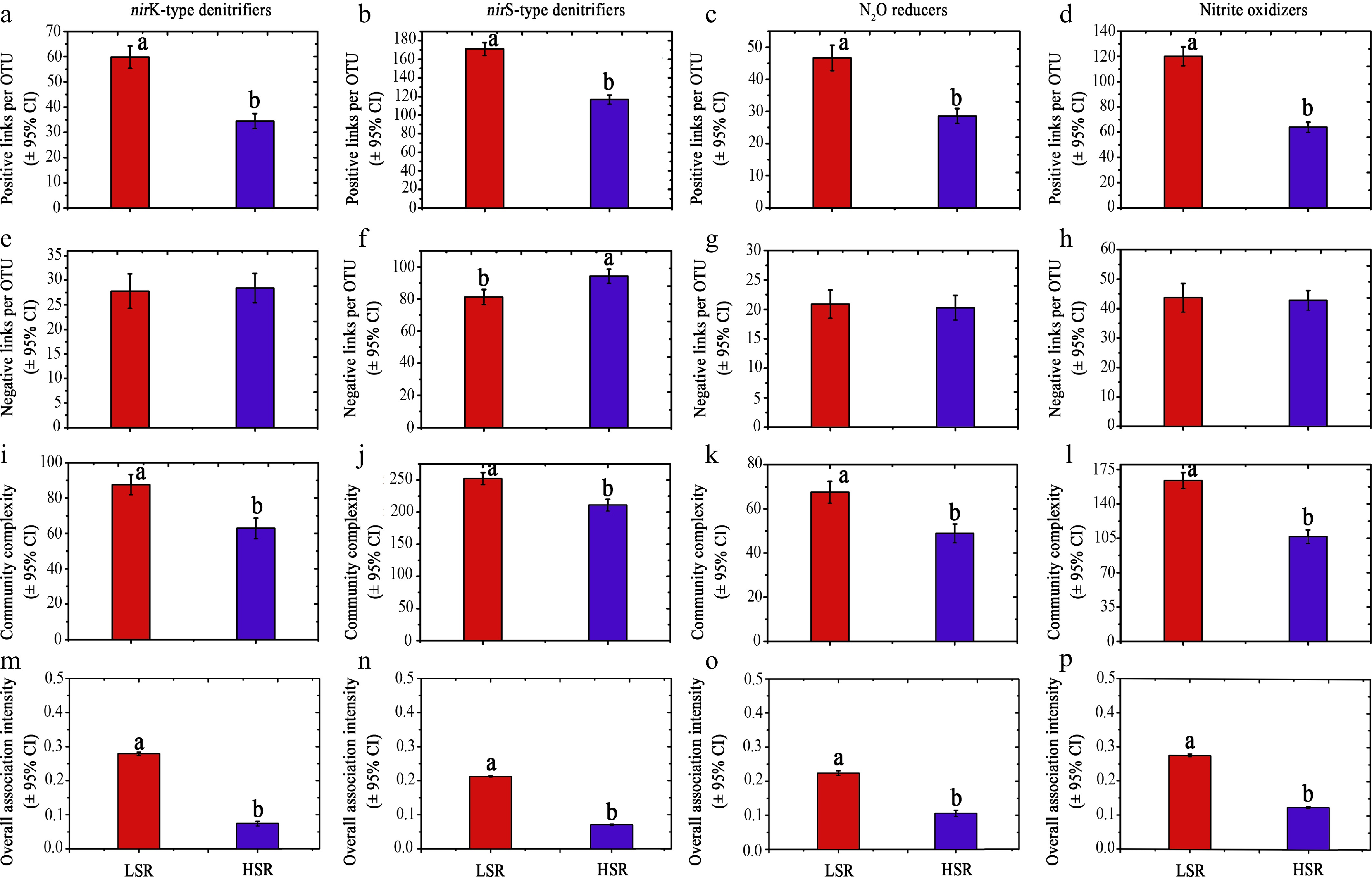
Figure 3.
The biotic associations and complexity for different N-transforming communities at low species richness (LSR) and high species richness (HSR).
-
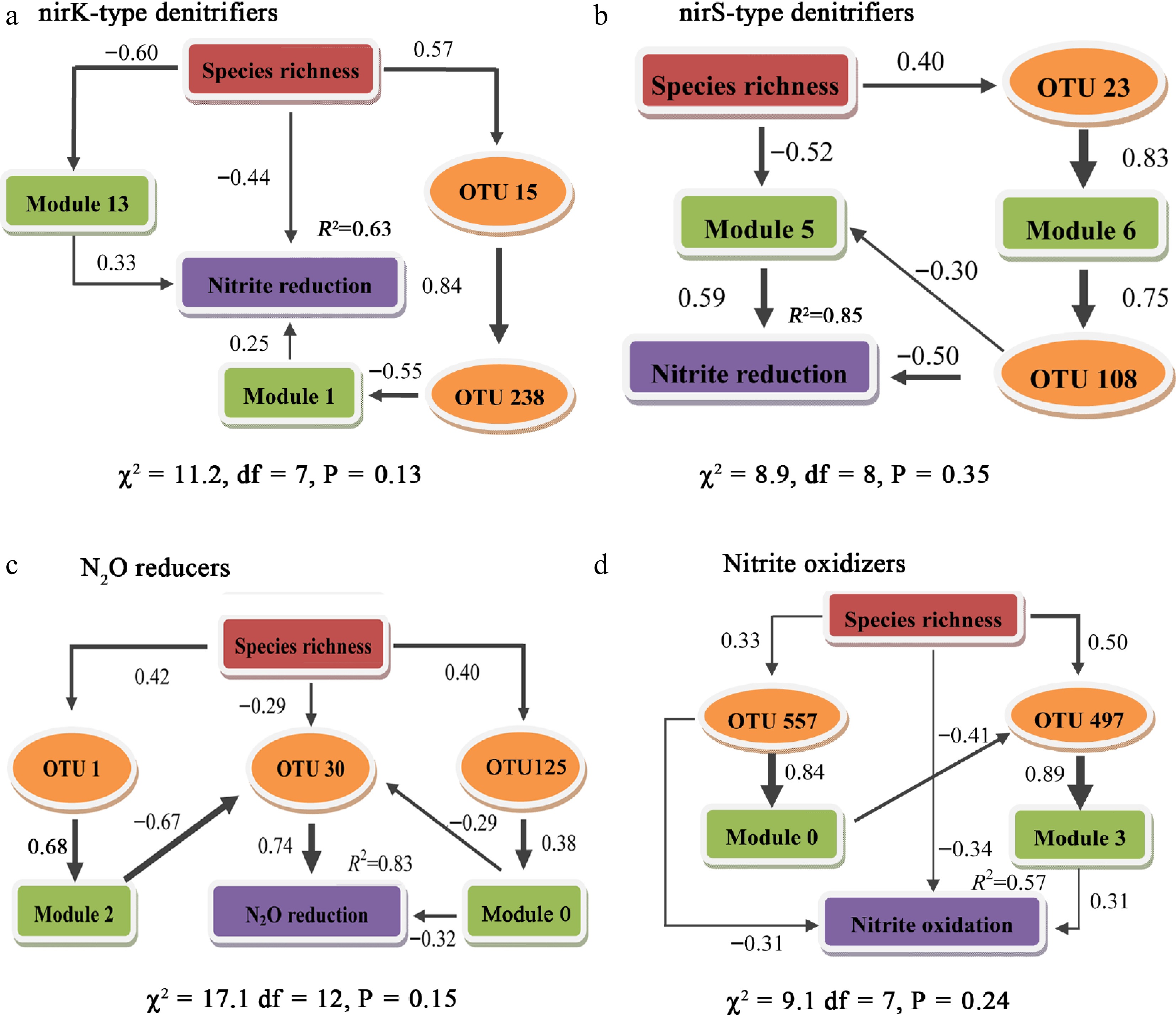
Figure 4.
Structural equation modeling to determine the direct and indirect effects using keystone species and dominant modules of species richness on N-transforming potential.
-
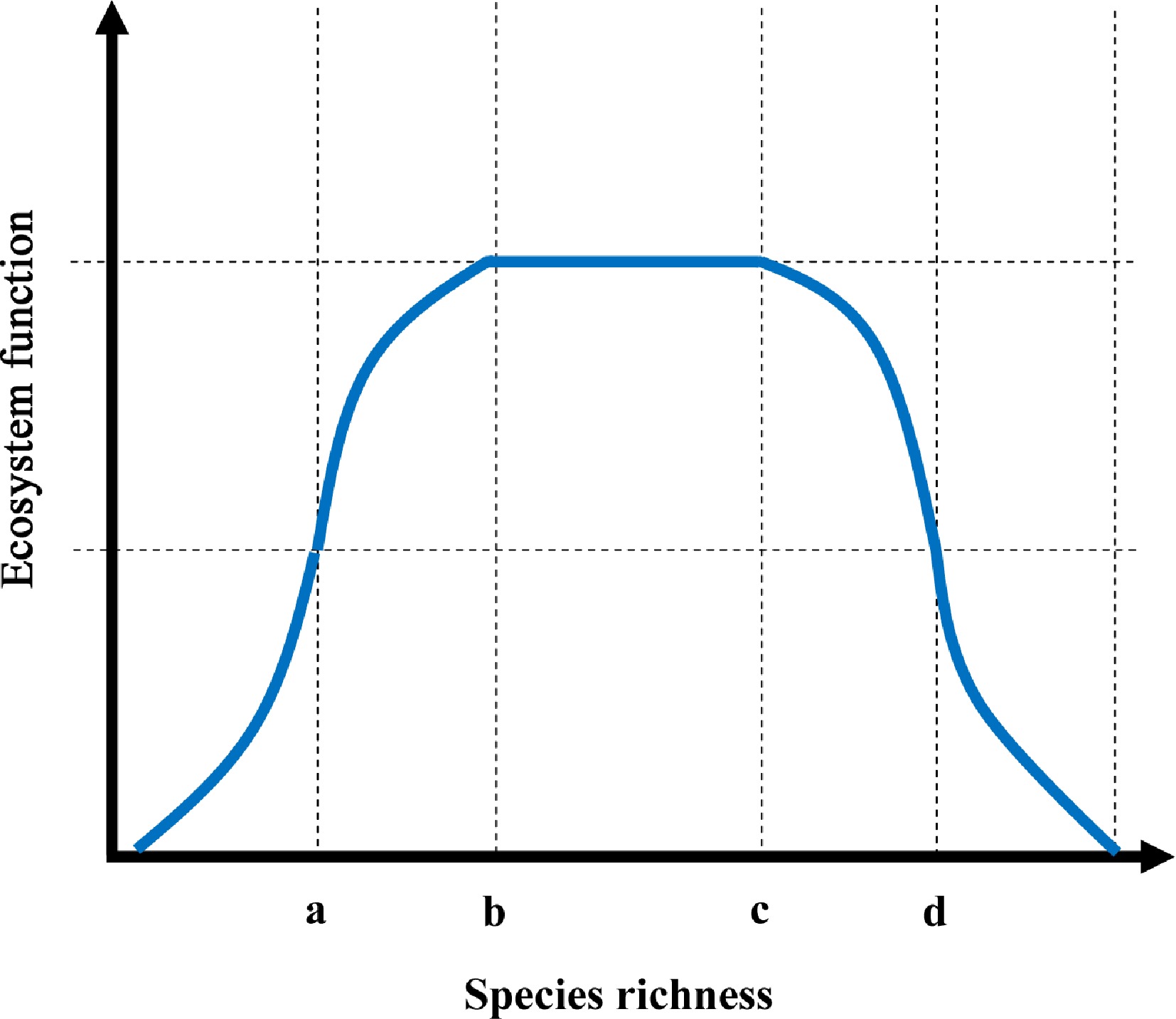
Figure 5.
A bell-shaped conceptual model for determining the relationship between species richness and ecosystem functioning.
-
Keystone taxa Roles Regressions (SR vs RA) Regressions (RA vs function) Importance (p value) Taxonomy nirK-type nitrite reducers OTU15 Module hub y = 0.005x − 2.309, p < 0.01 y = −0.476x + 4.006, p < 0.01 19.15 (0.05) Pannonibacter phragmitetus OTU173 Module hub y = −7.026 * 10−5x + 0.069, p < 0.05 y = 0.004x − 0.010, p = 0.20 11.17 (0.13) Devosia sp. GSM-205 OTU44 Connector y = 0.001x − 0.249, p < 0.05 y = −0.080x + 0.684, p < 0.01 17.30 (0.09) Bacterium clone_B-MYnirK1 OTU238 Connector y = 2.000 * 10−4x − 0.145, p < 0.01 y = −0.028x + 0.192, p < 0.01 33.73 (0.01) Chelativorans sp. BNC1 nirS-type nitrite reducers OTU2 Module hub y = 3.000 * 10−4x + 0.689, p = 0.53 y = −0.219x + 3.204, p < 0.01 28.70 (0.01) Rubrivivax gelatinosus OTU8 Module hub y = 4.000 * 10−4x + 0.092, p = 0.43 y = −0.057x + 1.570, p = 0.42 22.95 (0.02) Rubrivivax gelatinosus OTU12 Module hub y = 0.001x + 0.413, p = 0.39 y = −0.234x + 3.647, p < 0.01 26.43 (0.01) Uncultured bacterium OTU47 Module hub y = 3.000 * 10−4x − 0.208, p = 0.05 y = −0.037x + 0.722, p < 0.05 18.81 (0.03) Azospira sp. NC3H-14 OTU385 Module hub y = 4.618 * 10−5x − 0.074, p = 0.11 y = −0.009x + 0.113, p < 0.05 11.34 (0.10) Rhodocyclales bacterium OTU546 Module hub y = 2.000 * 10−4x − 0.146, p < 0.05 y = −0.035 + 0.650, p < 0.05 24.07 (0.03) Bacterium NirSIsoEc.72 OTU23 Connector y = 5.000 * 10−4x − 0.596, p < 0.05 y = −0.106x + 1.429, p < 0.01 25.86 (0.01) Uncultured bacterium OTU108 Connector y = 3.306 * 10−5x − 0.031, p = 0.09 y = −0.011x + 0.141, p < 0.01 39.37 (0.01) Rubrivivax gelatinosus N2O reducers OTU1 Module hub y = 0.011x − 3.112, p < 0.05 y = −0.313x + 6.878, p = 0.07 30.39 (0.03) Uncultured bacterium OTU30 Module hub y = −0.005x + 3.926, p < 0.01 y = 0.245x − 1.766, p < 0.01 31.31 (0.01) Herbaspirillum sp. TSO26-2 OTU67 Module hub y = −0.001x + 1.058, p < 0.01 y = 0.078x − 0.531, p < 0.01 18.62 (0.07) Massilia sp. TSO8 OTU261 Module hub y = −0.001x + 0.407, p < 0.01 y = 0.029x − 0.204, p < 0.01 26.78 (0.02) Azoarcus sp. CIB OTU109 Connector y = 3.000 * 10−4x − 0.148, p = 0.07 y = −0.012x + 0.166, p = 0.06 7.10 (0.26) Thauera humireducens OTU125 Connector y = 0.001x − 0.739, p < 0.05 y = −0.052x + 0.711, p < 0.05 30.97 (0.01) Uncultured bacterium Nitrite oxidizers OTU423 Module hub y = 0.012x − 9.331, p = 0.51 y = −0.389x + 9.769, p < 0.05 12.29 (0.13) Nitrospira calida OTU434 Module hub y = 0.007x − 4.520, p = 0.15 y = −0.090 + 3.743, p = 0.45 23.13 (0.01) Nitrospira calida OTU481 Module hub y = 0.005x − 4.109, p = 0.06 y = 0.003x + 1.382, p = 0.97 9.66 (0.19) Nitrospira moscoviensis OTU497 Module hub y = −0.009x + 10.065, p < 0.01 y = 0.193x − 2.490, p < 0.01 24.62 (0.01) Nitrospira moscoviensis OTU557 Module hub y = 0.002x − 1.758, p = 0.07 y = -0.091x + 2.022, p < 0.01 25.96 (0.01) Nitrospira moscoviensis OTU505 Connector y = −0.005x + 5.474, p < 0.05 y = 0.041x − 0.288, p = 0.48 4.20 (0.37) Nitrospira bockiana Values in bold indicate significant differences at alpha =0.05 level. Table 1.
Keystone taxa with significant changes identified based on network analysis.
Figures
(5)
Tables
(1)