-
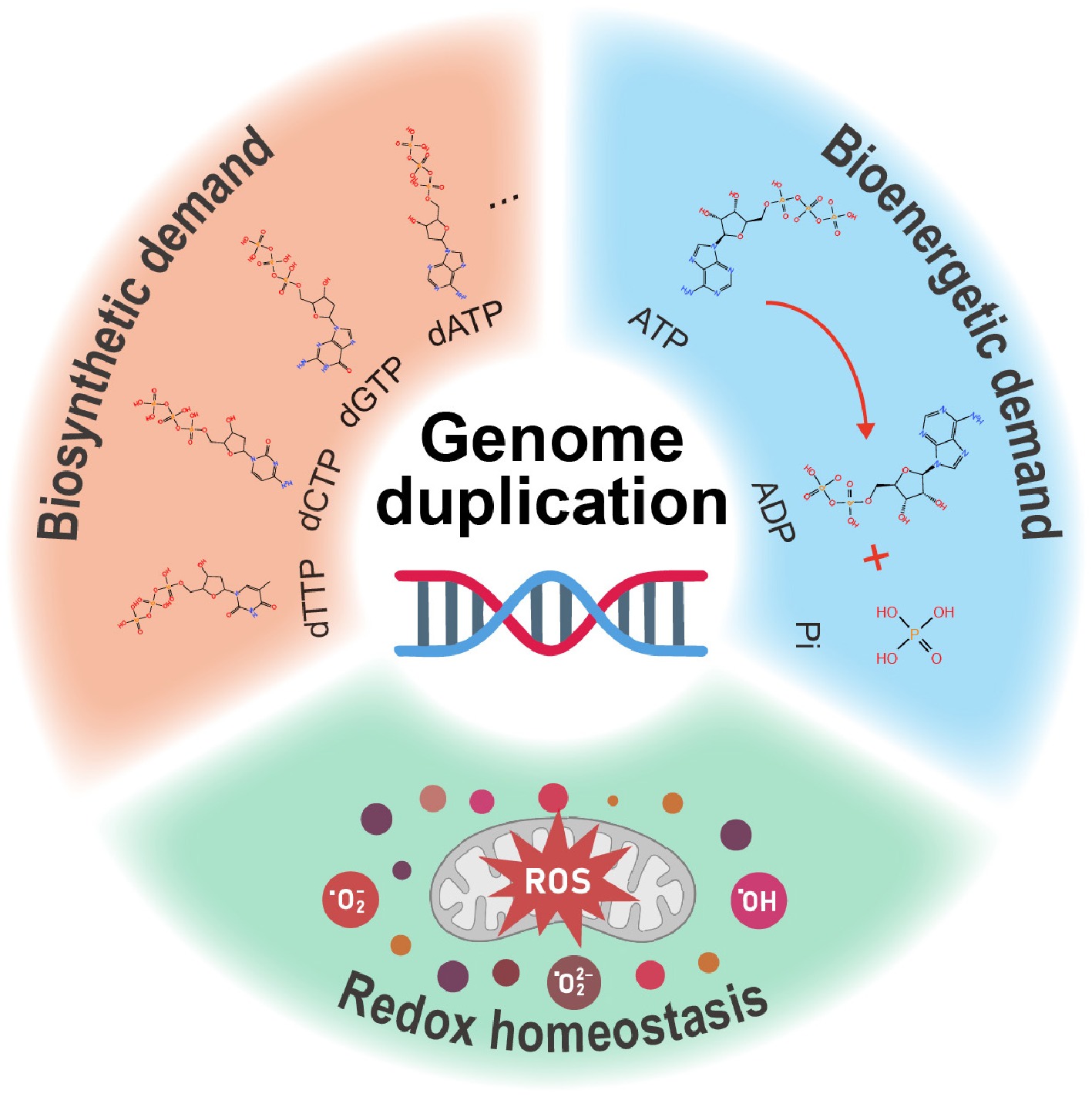
Figure 1.
DNA replication is coordinated with cell metabolism. (1) Biosynthetic demand: the biosynthesis of macromolecules is important for DNA replication. In particular, dNTP, including dTTP, dCTP, dGTP, and dATP, which serve as basic building blocks, are much in demand; (2) Bioenergetic demand: energy supply is also indispensable for DNA replication. For instance, changes in ATP level can affect DNA replication; (3) Redox homeostasis: hydrogen peroxide (H2O2), hydroxyl radical (·OH), and superoxide anion (·O2−) that are the main resources of ROS can influence DNA replication in different manners.
-
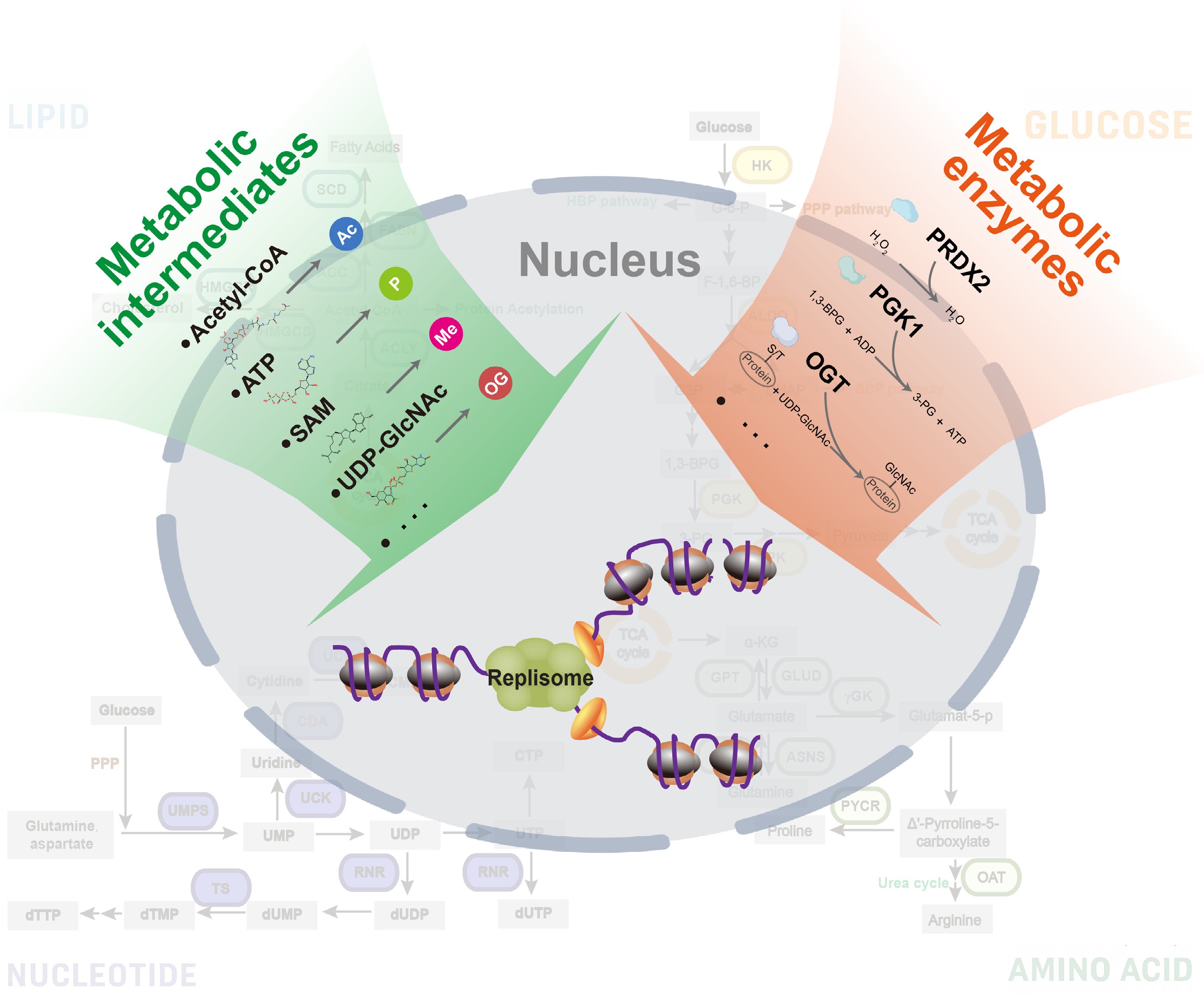
Figure 2.
Metabolic regulation of DNA replication. (1) Metabolic intermediates: small molecules, such as acetyl-CoA, ATP, SAM, and UDP-GlcNAc, function as donors and support the establishment of PTMs on key factors to regulate DNA replication; (2) Metabolic enzymes: classic enzymes that control metabolic reactions may enter the nucleus and regulate fork function/stability directly.
-
Protein Site(s) Function Ref. Replisome components MCM2-7 Unknown Promotes MCMs loading on chromatin [87] FEN1 S352 Disrupts FEN1 interaction with PCNA, causing replication defect and DNA damage [93] AND-1 S575, S893 Promotes HR to protect genome stability [94] Histones H4 S47 Directs DDK recruitment to promote replication origin activation [83] H2A S40 Associate with γH2AX to promote genome stability [84] DNA damage repair factors PLK1 T291 Required for proper chromosome segregation and thus important for the maintenance of genome stability [98] H2AX S139 Antagonizes phosphorylation of S139 on H2AX to restrain DNA damage signaling and protect genome integrity [90] Polη T457 Promotes TLS synthesis [99] MRE11 Unknown Promotes MRE11 recruitment on chromatin to protect genome integrity [100] RPA2 S4, S8 Antagonizes RPA2 S4/8 phosphorylation and impairs Chk1 activation [89] MCM, minichromosome maintenance; FEN1, flap endonuclease 1; AND-1, acidic nucleoplasmic DNA-binding protein 1; PLK1, polo-like kinase 1; Polη, DNA polymerase eta; MRE11, meiotic recombination 11; RPA, replication protein A; HR, homologous recombination. Table 1.
O-GlcNAcylation on factors that are potentially involved in the control of DNA replication.
-
Metabolic enzyme Function in DNA replication Ref. PGK1 Binds to CDC7 and convert ADP to ATP for origin activation [12] PRDX2 Associates with TIMELESS-TIPIN at the replication fork to adjust fork speed [11] OGT Enriches on chromatin in S phase and modifies H4 S47 to promote origin activation [83] ACO2 Knockdown of ACO2 compromises DNA synthesis [94] HK2 Knockdown of HK2 compromises DNA synthesis [94] GAPDH Knockdown of GAPDH compromises DNA synthesis [94] PDC Localizes in the nucleus and regulates S phase entry [97] H6PD Knockdown of H6PD impairs DNA synthesis [93] RPE Knockdown of RPE impairs DNA synthesis [93] PRPS1 Knockdown of PRPS1 impairs DNA synthesis [93] ACLY Regulates nuclear acetyl-CoA concentration thereby regulating MCM2-7 and CDC45 transcription [101] FH Knockout of FH in S. cerevisiae leads to be sensitive to HU [95] IDH1/2 IDH1/2 mutants regulate DNA replication by contributing heterochromatin formation [96] PGK1, phosphoglycerate kinase 1; PRDX2, peroxiredoxin 2; OGT, O-GlcNAc transferase; ACO2, aconitate hydratase 2; HK2, hexokinase 2; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PDC, pyruvate dehydrogenase complex; H6PD, hexose-6-phosphate dehydrogenase; RPE, ribulose 5-phosphate epimerase; PRPS1, ribose-phosphate pyrophosphokinase 1; ACLY, ATP-citrate lyase; FH, fumarate hydratase; IDH, isocitrate dehydrogenase. Table 2.
Metabolic enzymes potentially involved in the regulation of DNA replication.
Figures
(2)
Tables
(2)