-
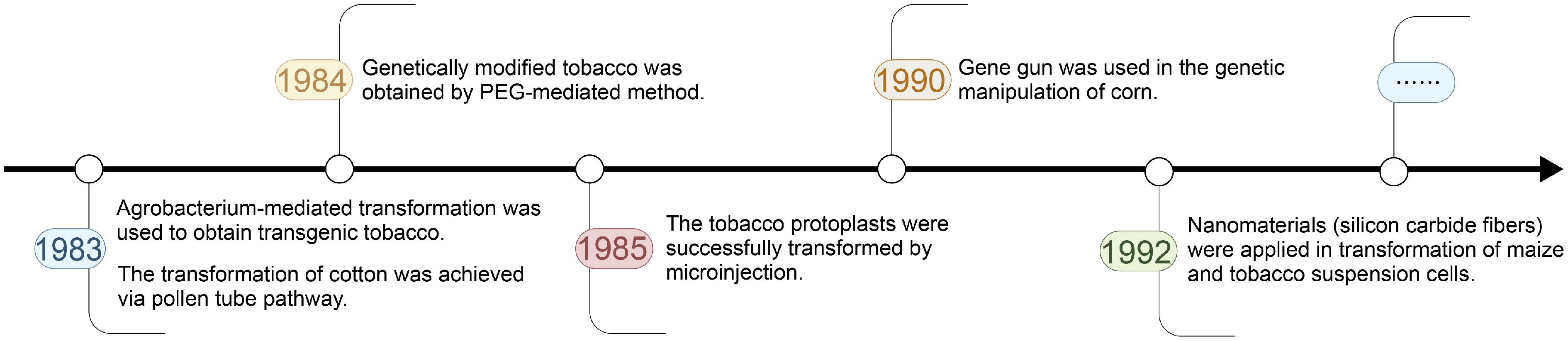
Figure 1.
The timeline of the development of plant genetic transformation technologies.
-
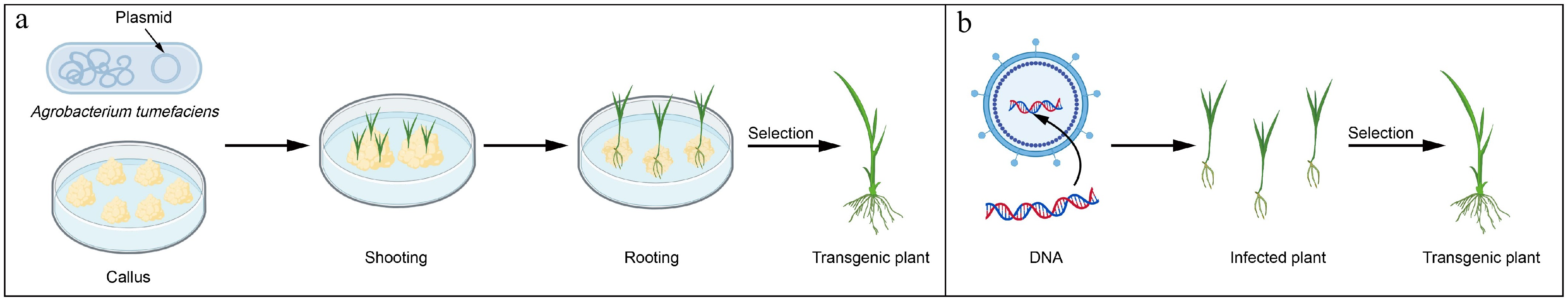
Figure 2.
Schematic diagram of biologically mediated transformation.
-
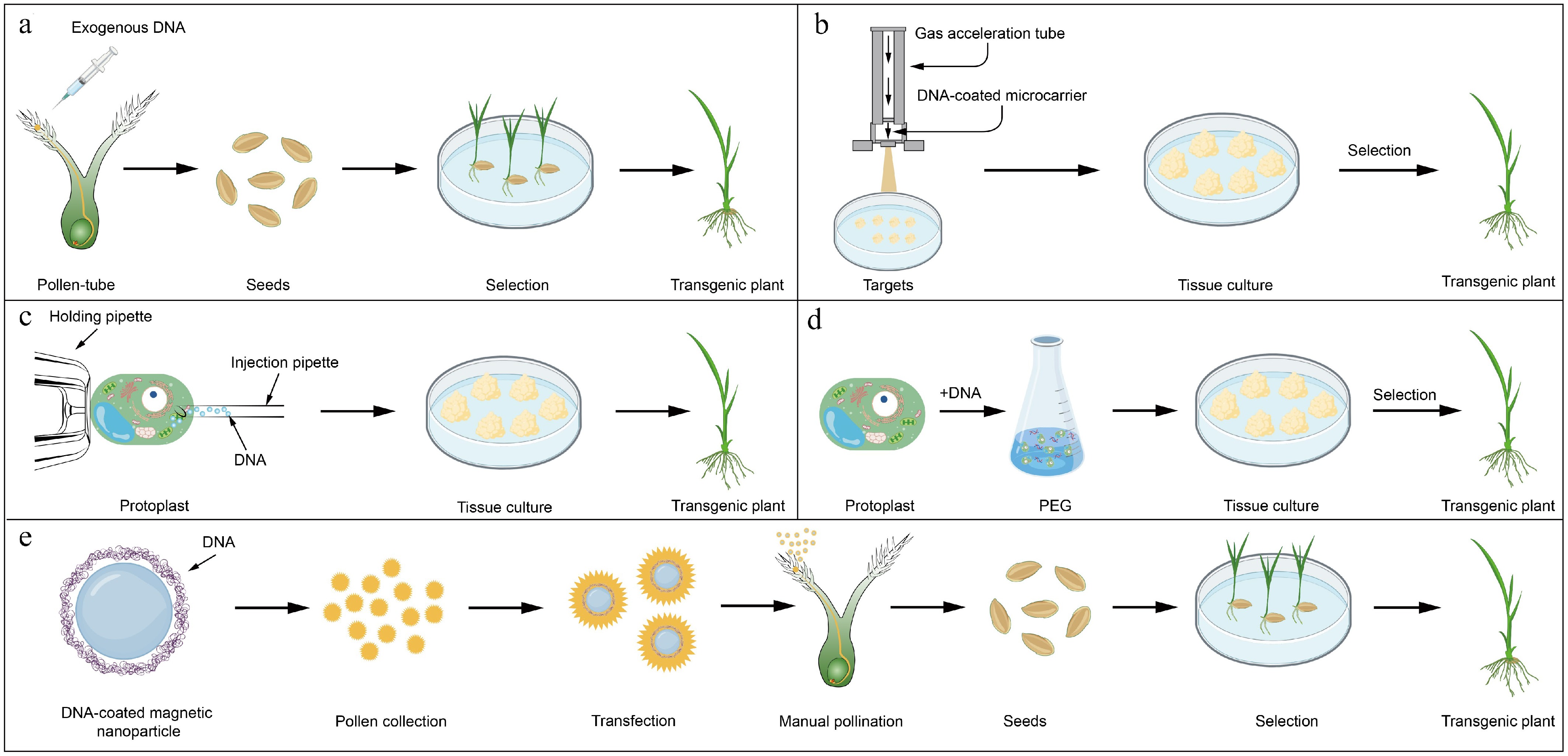
Figure 3.
Schematic diagram of non-biologically mediated transformation.
-
Genes Species Delivery context Expression strategy Transformation effects Ref. WUS, BBM Maize Agrobacterium Constitutive expression Significantly increases transformation efficiency and reduces genotype dependence [17] Rice Sorghum Maize Agrobacterium Specific expression Transformation frequency varied between inbred lines with averages ranging from 9% for one ear from B73 to 224% for one ear from PHH5G [18] Maize Agrobacterium Specific expression The transformation frequency in the T0 generation is 29%–69% [19] WUS Sorghum Agrobacterium Specific expression Wus2 enhances both transformation efficiency and CRISPR editing frequency in sorghum [20] BBM Apple Agrobacterium Constitutive expression The transformation efficiency was 3.68%–28.15% [21] WIND1 Arabidopsis Agrobacterium Constitutive expression Greatly enhances de novo shoot regeneration [22] Rapeseed GRF3-GIF1 Soybean Agrobacterium Constitutive expression The average transformation efficiency with the GRF3-GIF1 chimera was 2.74-fold higher than for the empty vector control [23] GRF5 Sugar beet Agrobacterium Constitutive expression Transformation efficiency was increased 6-fold [24] Canola Promoted the proliferation of transgenic callus cells Soybean Promoted the production of transgenic shoots Sunflower Promoted the production of transgenic shoots Maize Transformation frequencies increase by three times for inbred line A188 TaDOF5.6, TaDOF3.4 Wheat Agrobacterium Constitutive expression The transformation efficiency was improved by about 17%–34% [25] WOX5 Wheat Agrobacterium Constitutive expression Successfully transformed 31 common wheat cultivars, overcoming genotype dependency [26] Triticum monococcum PI428182 showed a significant increase in transformation efficiency to 94.5% ± 8.2% Rye Transformation efficiency of 7.8% in Lanzhou Heimai Triticale Transformation efficiency of 16.4%–53.3% in Linfen45, ZS3297, ZS1257, and ZS3224 Barley Transformation efficiency of 10.4%–88.4% in barley Maize Transformation efficiency of 19.2%–38.4% in B73 and A188 AIL5 Apple Agrobacterium Constitutive expression Significantly improved the regeneration efficiency of adventitious shoot [27] Table 1.
Developmental regulators applied in plant genetic transformation.
-
Methods Details Species Ref. Modification of Agrobacterium Introducing helper plasmids Maize [28] Modifying Agrobacterium tumefaciens with T3SS Arabidopsis thaliana, Nicotiana benthamiana, Wheat, Alfalfa, Switchgrass [29] Expressing the DC3000 EF-Tu variants Arabidopsis thaliana, Camelina sativa [30] Improvement of nanomaterials Polyethyleneimine (PEI)-modified single-walled carbon nanotubes (PEI-SWCNTs) Nicotiana benthamiana, Arugula, Wheat, Cotton [31] Modified carbon dots (MCDs) Wheat [34] Ferroferric oxide (Fe₃O₄) magnetic nanoparticles (MNPs) Maize [32] Mesoporous silica nanoparticles (MSNs) Oryza alta [33] Increase of plant cell permeability Surfactants Wheat [35] Sonication Soybean, Rubber tree [16,36] Table 2.
Other methods to improve the efficiency of plant genetic transformation.
Figures
(3)
Tables
(2)