-
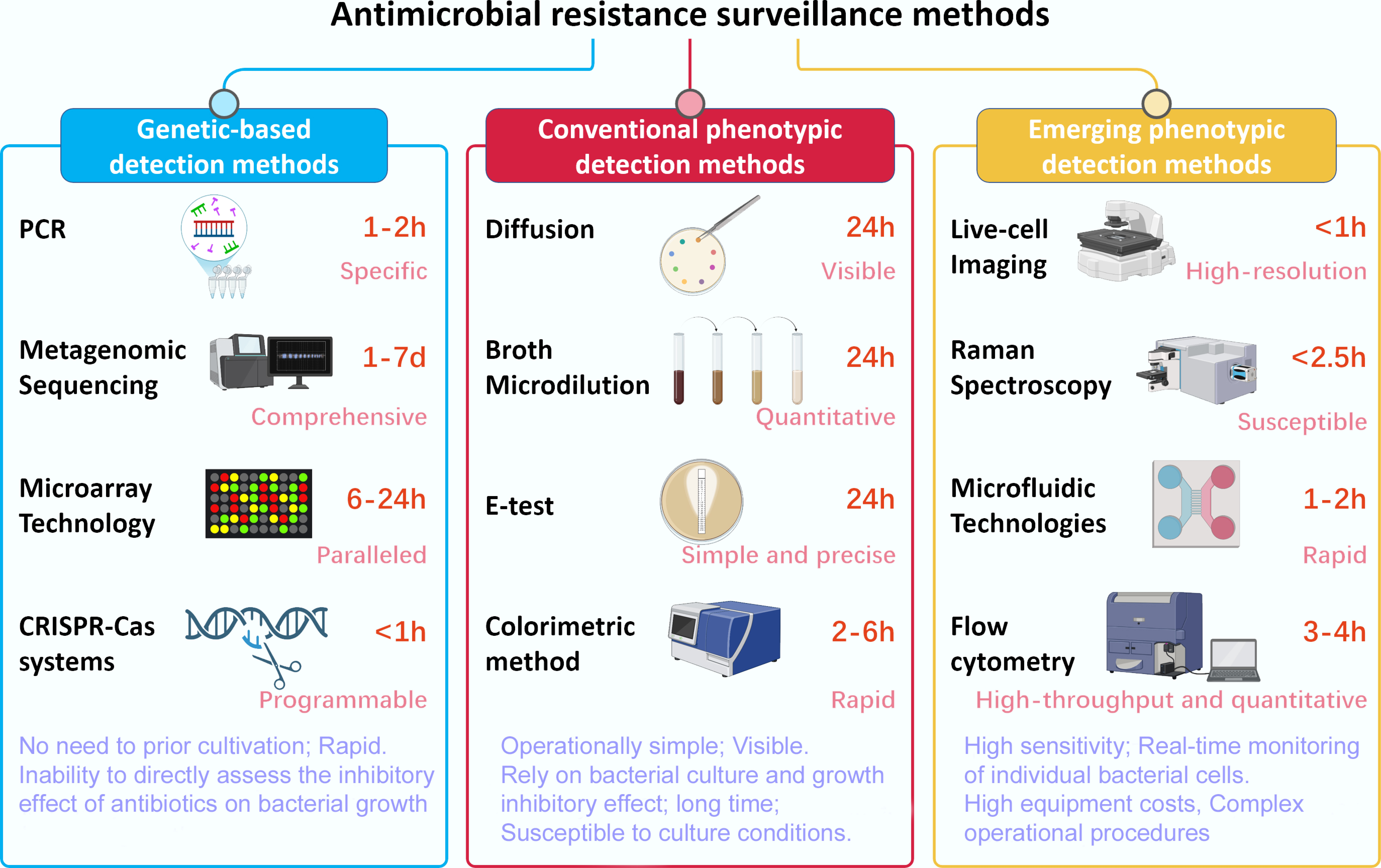
Figure 1.
The typical antimicrobial resistance surveillance methods include genetic-based, conventional phenotypic, and emerging phenotypic methods.
-
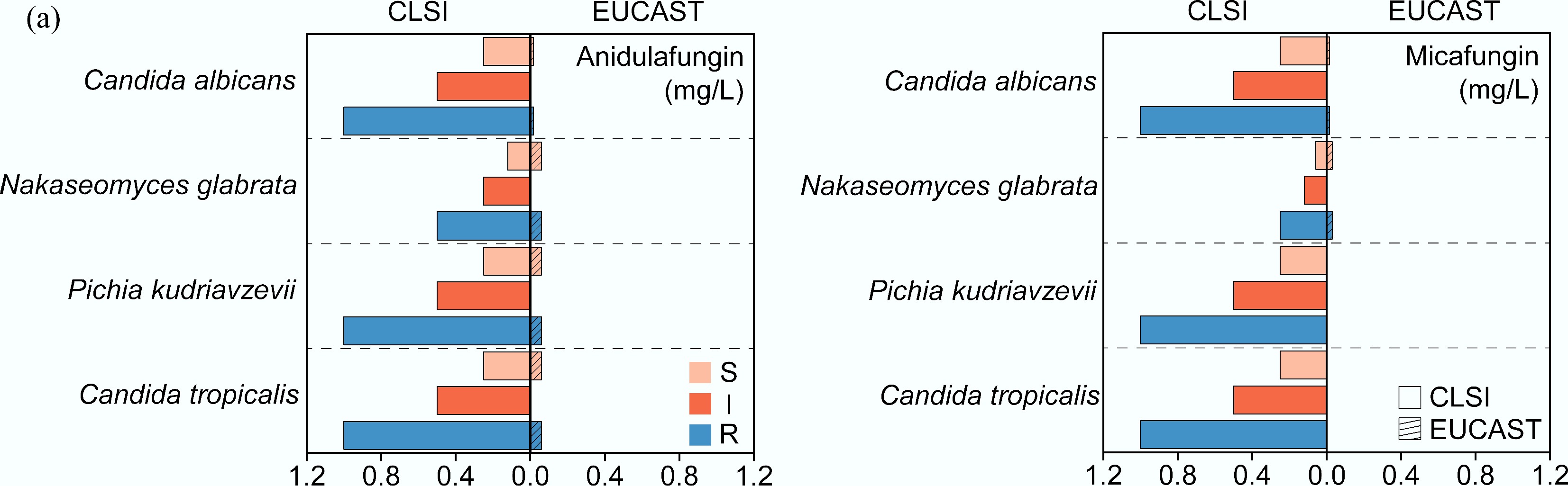
Figure 2.
Comparison of MIC breakpoints for (a) anidulafungin and (b) micafungin between CLSI and EUCAST. S: Susceptible, I: Intermediate, R: Resistant[91].
-
Technology Main principle Advantages and disadvantages Ref. PCR Detect antimicrobial resistance by amplifying resistance genes. Advantages: high sensitivity; detected rapidly (within 1–2 h)
Disadvantages: detecting the specific resistance genes; cannot assess antibiotic susceptibility[21,53−55] Metagenomic sequencing Analyze the total microbial DNA in a sample through high-throughput sequencing. Advantages: comprehensive without prior cultivation
Disadvantages: standardization challenges currently limited their clinical adoption[22,56,57] DNA microarray Immobilize specific probes on a chip and detect resistance genes in the sample through hybridization reactions. Advantages: high throughput (able to detect multiple resistance genes simultaneously); rapidness (suitable for clinical application)
Disadvantages: high cost; complex operation; and the risk of false positives (sample contamination)[40,58] CRISPR-Cas Utilize the RNA-guided nucleic acid recognition capability to detected ARGs with single-base resolution. Advantages: rapid (< 1 h), instrument-free detection suitable for point-of-care applications
Disadvantages: currently lack standardized protocols for widespread clinical implementation[42,43] Disk diffusion method The susceptibility is determined by the inhibition zone formed by the diffusion of antibiotics in the culture medium. Advantages: easy to perform; cost-effective; can test the susceptibility to multiple antibiotics simultaneously; suitable for routine clinical laboratories with limited resources
Disadvantages: partially automated; insufficient data support or poor detection performance for some bacteria; susceptible to environmental factors[23−26] Broth microdilution method The MIC is determined by the inhibition of bacterial growth at different concentrations of antibiotics. Advantages: accurately determine the MIC; has a high degree of standardization
Disadvantages: reagent requirements; high cost; complex operation; risk of false positives; risk of cross-contamination; the inability to distinguish between live and dead bacteria[37,38,45] E-test The MIC is determined by measuring the elliptical zone of bacterial growth inhibition created by a plastic strip containing a gradient concentration of antibiotics. Advantages: easy to operate; accurate in results; highly sensitive; suitable for a variety of bacteria, including slow-growing bacteria such as Helicobacter pylori
Disadvantages: not accurate enough for certain antibiotics (such as penicillin and ciprofloxacin); costly; has strict requirements for operating conditions[27−29] Colorimetric method Assess antibiotic susceptibility by detecting color changes caused by bacterial metabolism. Advantages: simple to use; low-cost; compatible with the standard microdilution method; does not require complex equipment
Disadvantages: low specificity; easily interfered with by other redox substances; long detection time (4–6 h)[46,47] Forward light scattering Detect changes in bacterial growth using laser scattering technology. Advantages: high throughput; rapid detection
Disadvantages: cannot distinguish between live and dead bacteria; high background noise[48,49] Real-time microscopy Conduct rapid detection using real-time microscopic imaging and fluorescence in situ hybridization (FISH) technology. Advantages: real-time monitoring (4–9 h); high degree of automation (the entire process from sample pretreatment to result analysis is automated)
Disadvantages: complex (requiring a high-precision microscope and an automated system); low throughput (only one sample can be processed at a time)[59−62] Live-cell imaging Monitor the growth and division of individual bacteria using microfluidic channels and microscopic imaging techniques. Advantages: rapid; high-throughput
Disadvantages: complex equipment and high cost equipment (high-resolution microscope and microfluidic system); high maintenance costs[33,62] Isothermal microcalorimetry Assess antibiotic susceptibility by detecting changes in heat produced by bacterial metabolism. Advantages: high sensitivity; free from matrix interference
Disadvantages: complex; data interpretation is relatively complicated[51,63] Mass-sensitive technology The technology based on microcantilevers assesses antibiotic susceptibility by detecting changes in bacterial mass. Advantages: high sensitivity (able to detect changes in individual bacteria); rapid (within 3–4 h)
Disadvantages: complex and high cost equipment (high-precision microcantilevers and vacuum systems); high maintenance costs[64,65] Electrical Assess antibiotic susceptibility by detecting changes in electrical properties such as current and voltage. Advantages: rapid (1–3 h); good portability
Disadvantages: low specificity; high background noise.[50,51] Motion tracking Assess antibiotic susceptibility by monitoring changes in bacterial movement using optical tracking technology. Advantages: rapid (within 30 min to 2 h); high sensitivity (detect changes at the single-cell level)
Disadvantages: complex and high cost equipment (high-precision microscopes and image processing algorithms); high maintenance costs[33,66] Raman spectroscopy Assess antibiotic susceptibility by detecting the Raman spectra of bacteria. Advantages: high sensitivity; rapid (within 30 min to 2.5 h)
Disadvantages: complex equipment; high background noise[30−32] Laser tweezers raman spectroscopy Capture individual bacteria with laser tweezers and perform Raman spectroscopy analysis. Advantages: single-cell analysis capability; background signal elimination
Disadvantages: complex equipment and operation[67,68] Fast Raman-assisted antibiotic susceptibility test Detect single bacterial metabolic activity in the presence of antibiotics, using Raman single-cell spectroscopy. Advantages: rapid (3 h) for urinary antibiotic susceptibility testing, enabling rapid clinical decision-making
Disadvantages: relies on sophisticated instrumentation and technical expertise[69,70] Fluorescence detection Detect the growth of bacteria by detecting metabolic activity. Advantages: high sensitivity (detect changes close to the single-cell level); rapid (within 3–4 h)
Disadvantages: high background noise; complex optical equipment and operation; poor performance on bacteria with low metabolic activity and the need for pure culture colonies[71−74] Microfluidic technologies Capture individual bacteria using microfluidic chips and conduct detection. Advantages: rapid (1–2 h); high throughput
Disadvantages: complex; strict limitations on sample volume[33,68] Flow cytometry Assess antibiotic susceptibility by detecting the scattered light and fluorescence of individual bacteria. Advantages: rapid detection; simultaneous analysis of multiple cell characteristics; high throughput; quantitative
Disadvantages: complex sample preparation; limited ability to analyze rare cells; high cost; complex equipment and operation[52,75] Table 1.
Comparison of antimicrobial resistance surveillance methods
-
Antibiotic Strain and the test number CLSI (%) EUCAST (%) S I R S I R Amoxicillin-clavulanate Escherichia coli (428) 55.6 24.5 19.9 47.7 − 52.3 Klebsiella pneumoniae (208) 67.3 10.6 22.1 64.4 − 35.6 Ciprofloxacin Escherichia coli (428) 50.5 3 46.5 31.3 9.6 59.1 Klebsiella pneumoniae (208) 72.6 13.5 13.9 47.6 14.9 37.5 Pseudomonas aeruginosa (78) 85.9 3.8 10.3 71.8 0 28.2 Gentamicin Escherichia coli (428) 58.4 0 41.6 55.1 2.8 42.1 Klebsiella pneumoniae (208) 76.4 0 23.6 73.6 2.9 23.6 Pseudomonas aeruginosa (78) 76.9 0 23.1 75.6 0 24.4 Ceftriaxone Escherichia coli (428) 41.8 0.2 57.9 40.4 1.4 58.2 Klebsiella pneumoniae (208) 68.8 0 31.3 64.9 3.8 31.3 The number of strains refers to the total count of bacterial isolates tested for their susceptibility to antibiotics[87−89]. A higher percentage in the S category indicates a greater number of susceptible isolates among the tested strains, while a higher percentage in the R category indicates a greater number of resistant isolates. S: Susceptible, I: Intermediate, R: Resistant. Table 2.
Susceptibility analysis of different antibiotics to bacteria based on CLSI and EUCAST
Figures
(2)
Tables
(2)