-
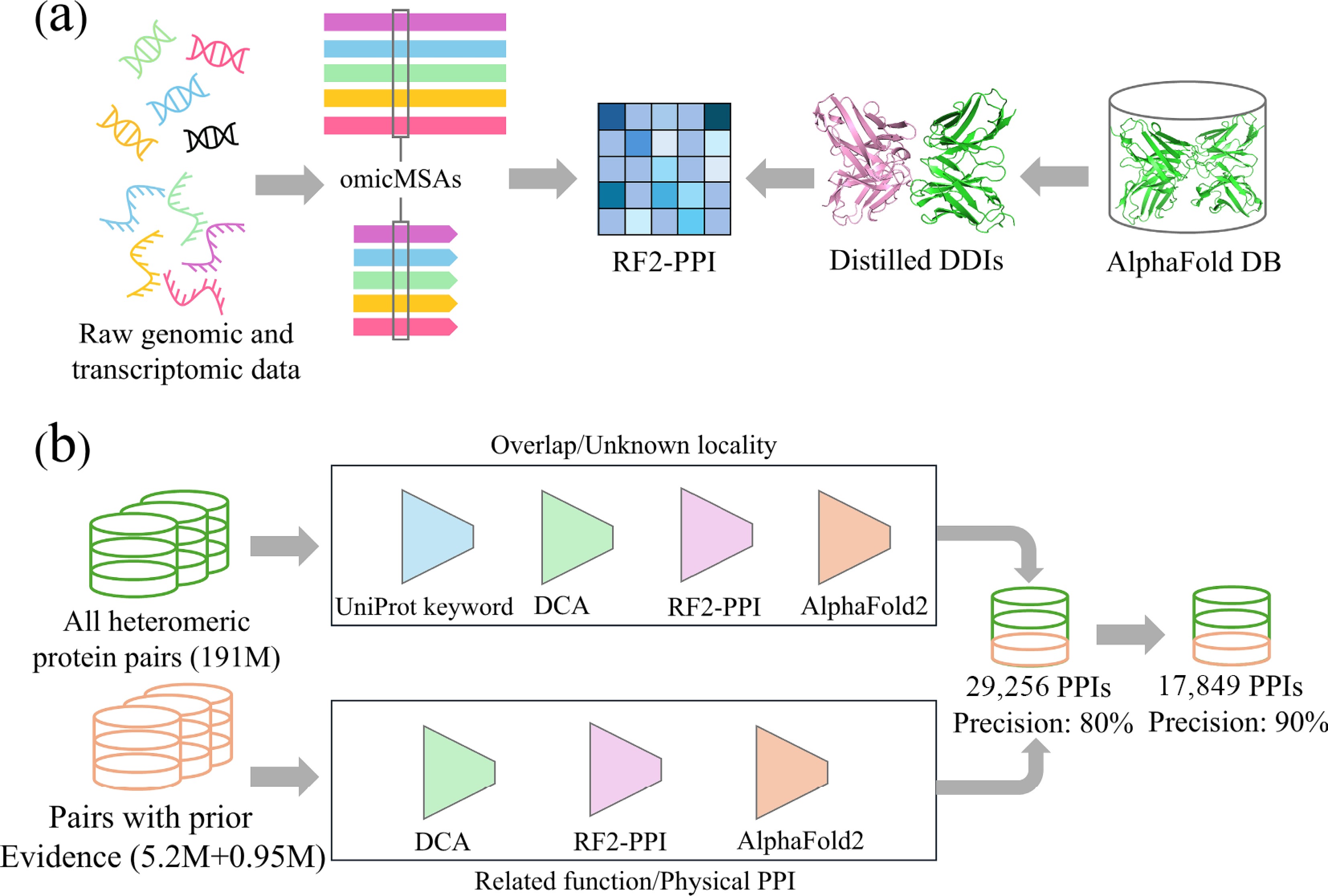
Figure 1.
Large-scale human PPI prediction analysis diagram based on deep learning. (a) Integration of sequence and structure information to train RF2-PPI. Raw genomic and transcriptomic data from diverse eukaryotes are processed to construct omicMSAs. In parallel, high-confidence intra-chain domain-domain interactions are distilled from AlphaFold DB monomer models. These sequence-based omicMSAs and structure-derived distilled DDIs are jointly used to train RF2-PPI. (b) Schematic overview of the two complementary screening pipelines used to predict human PPIs. For the de novo search, all heteromeric protein pairs formed by screened proteins are restricted to proteins with overlapping or unknown subcellular localization. They are then sequentially filtered by UniProt keyword-based co-localization, DCA, RF2-PPI scoring, and AlphaFold2 complex modelling. For the evidence-guided search, protein pairs with prior evidence are evaluated by DCA, RF2-PPI and AlphaFold2 using more permissive score thresholds.
-
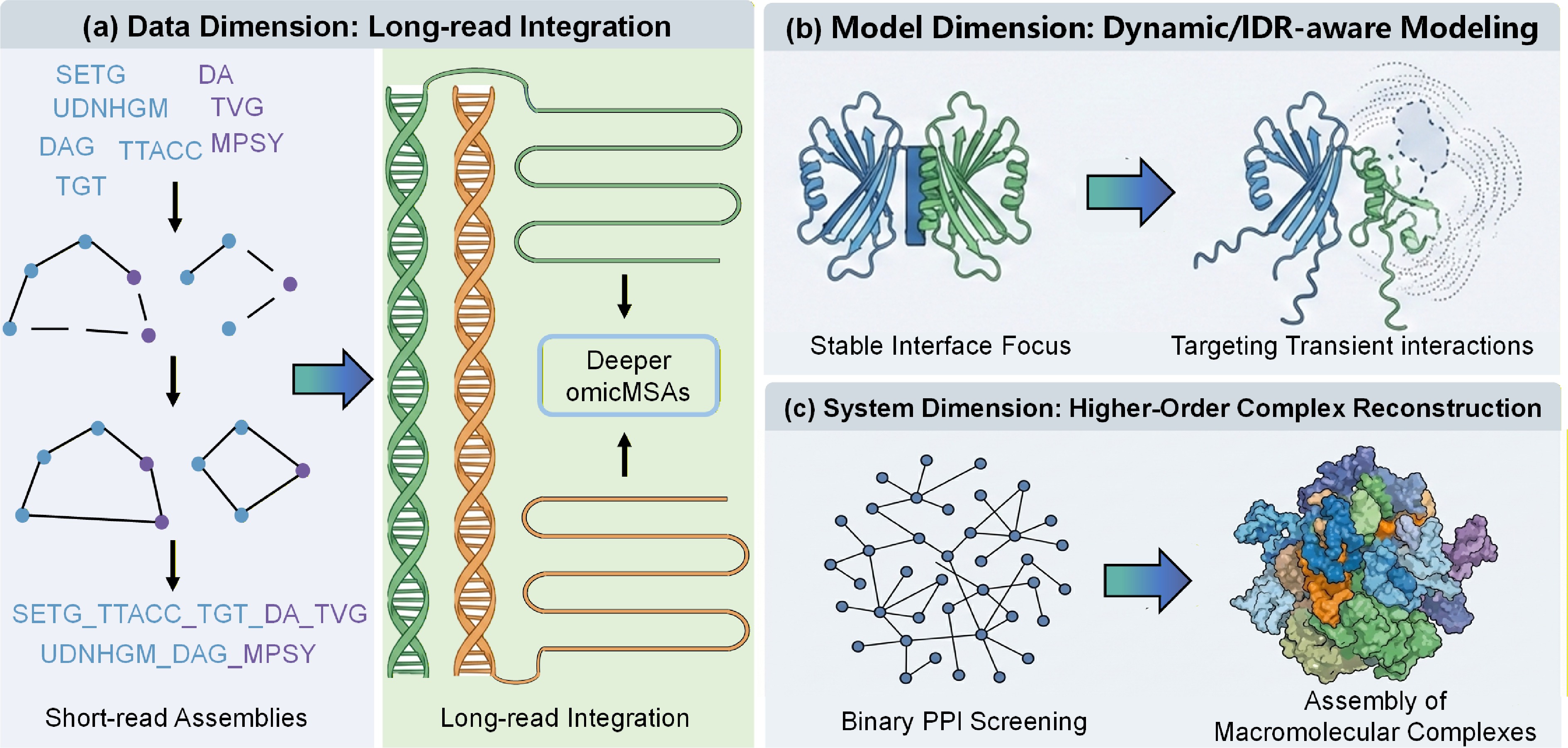
Figure 2.
A roadmap for next-generation interactome prediction. The figure illustrates three dimensions for extending the current framework. (a) Data Dimension, moving from short-read assemblies to long-read integration to resolve difficult eukaryotic genomes and maximize evolutionary signal depth. (b) Model Dimension, evolving from stable interface focus to dynamic/IDR-aware modeling to capture transient signaling interactions and disordered regions often missed by rigid-body docking. (c) System Dimension, scaling up from binary PPI screening to higher-order complex reconstruction, integrating binary predictions into graph-based assembly of large molecular machines.
Figures
(2)
Tables
(0)