-

Figure 1.
Fruit profiles at defined peach developmental stages (scale bar = 1 cm).
-
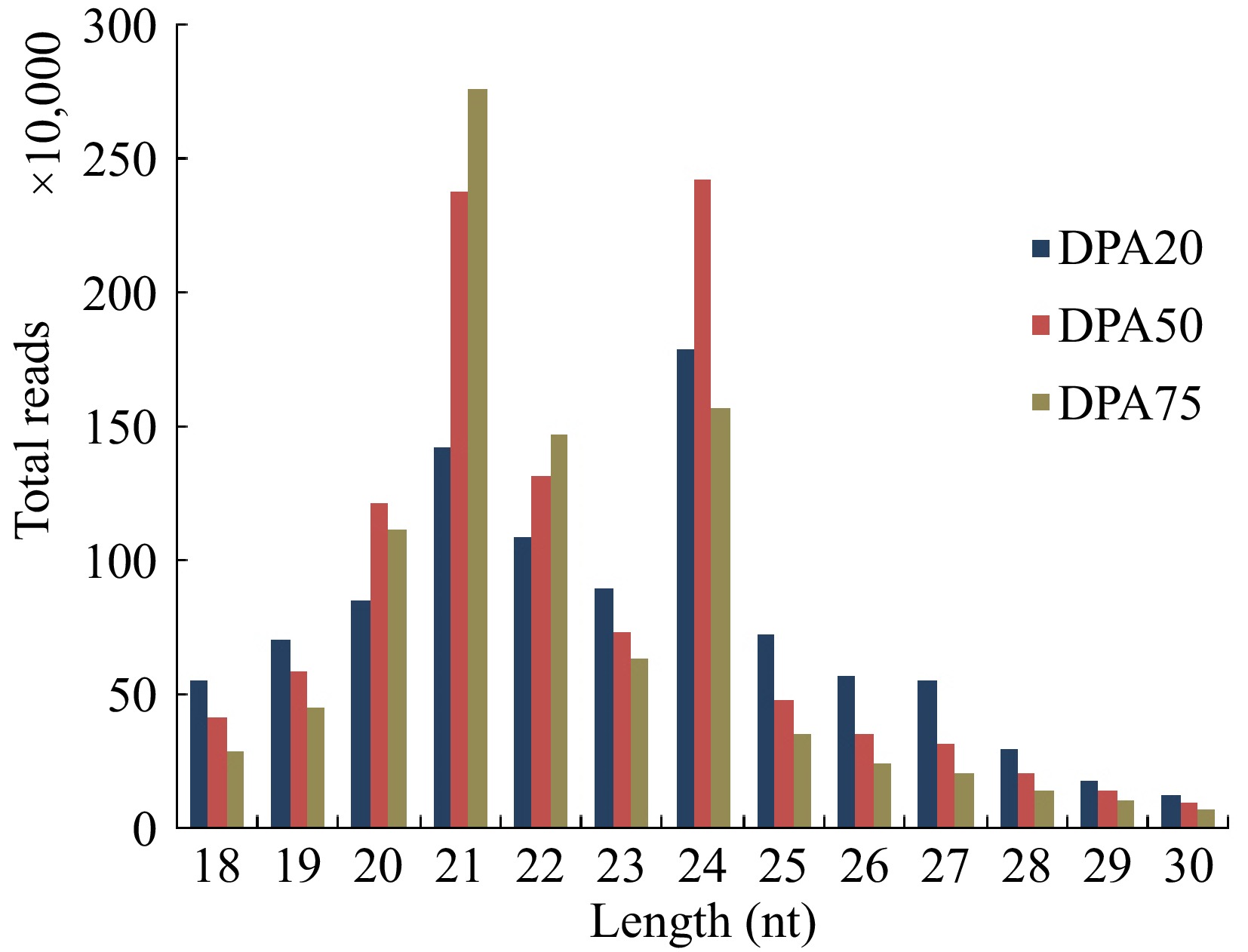
Figure 2.
Length distributions of sRNAs of peaches in the libraries at 20 d post anthesis (DPA20), DPA50, and DPA75.
-
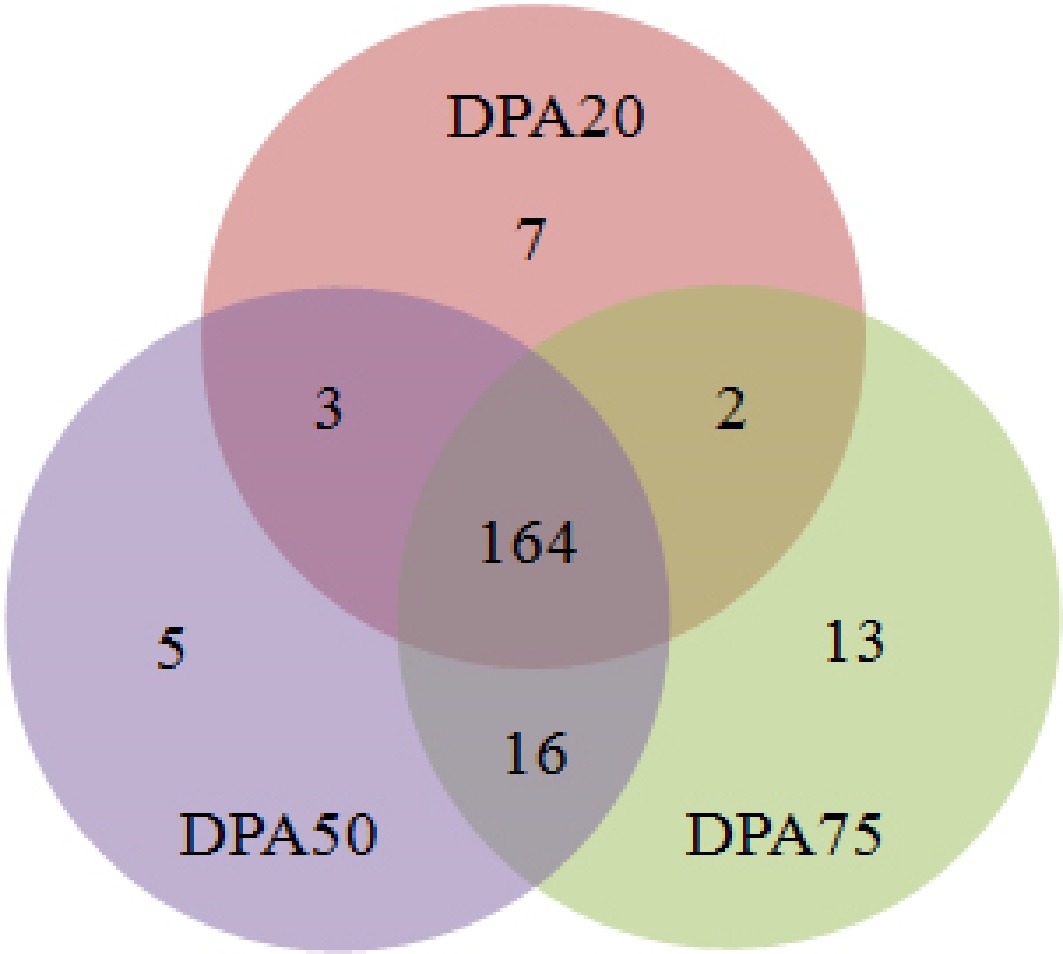
Figure 3.
Venn diagram of peach miRNAs among these libraries at DPA20, DPA50, and DPA75.
-
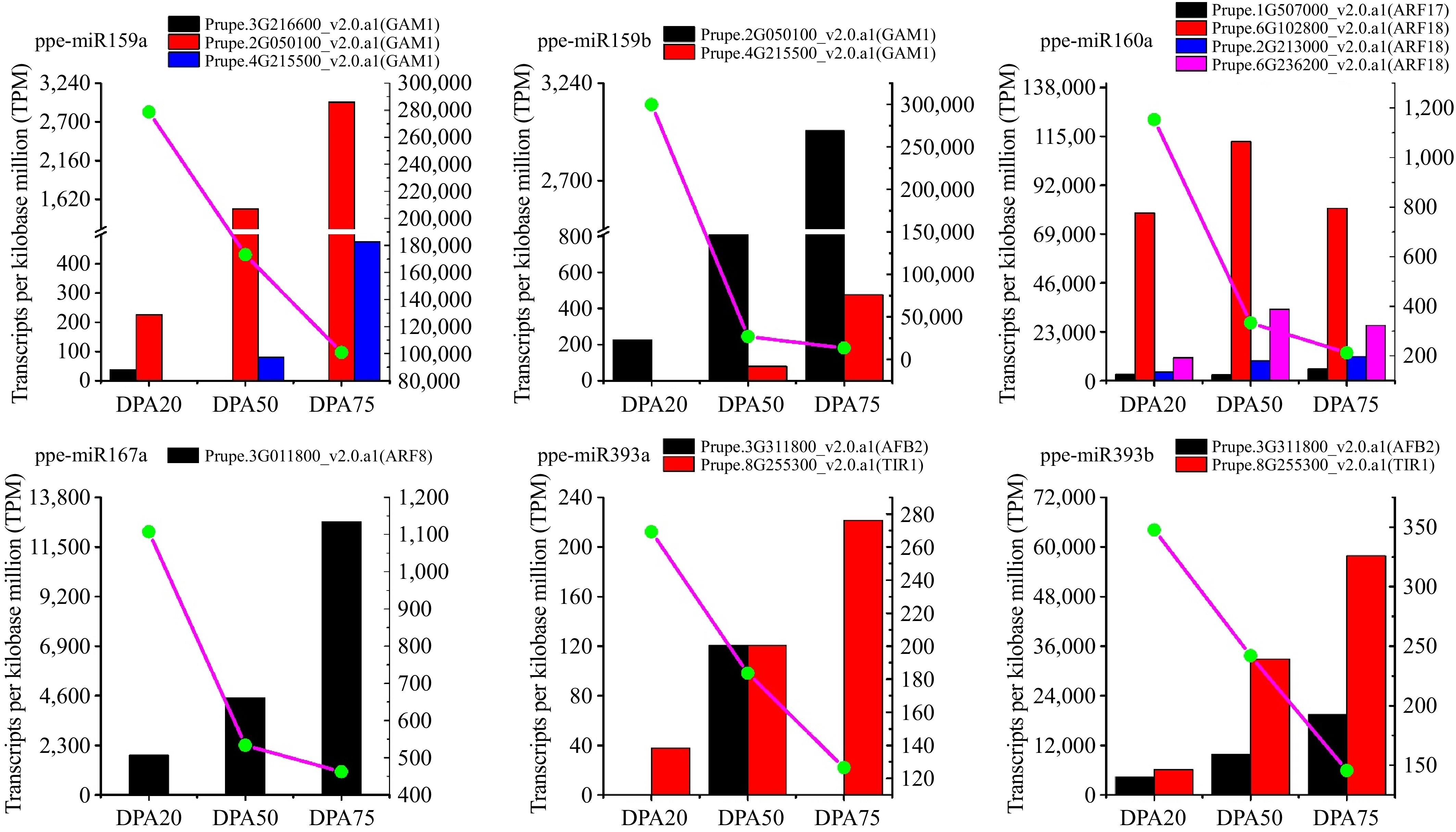
Figure 4.
Abundance of miRNA and its target genes at different peach developmental stages. Normalized sequenced read abundance, height = transcripts per kilobase million reads (TPM) on the y-axis. The histogram represents the abundance of miRNA target genes and the dot linked by a line represent the abundance of miRNA.
-
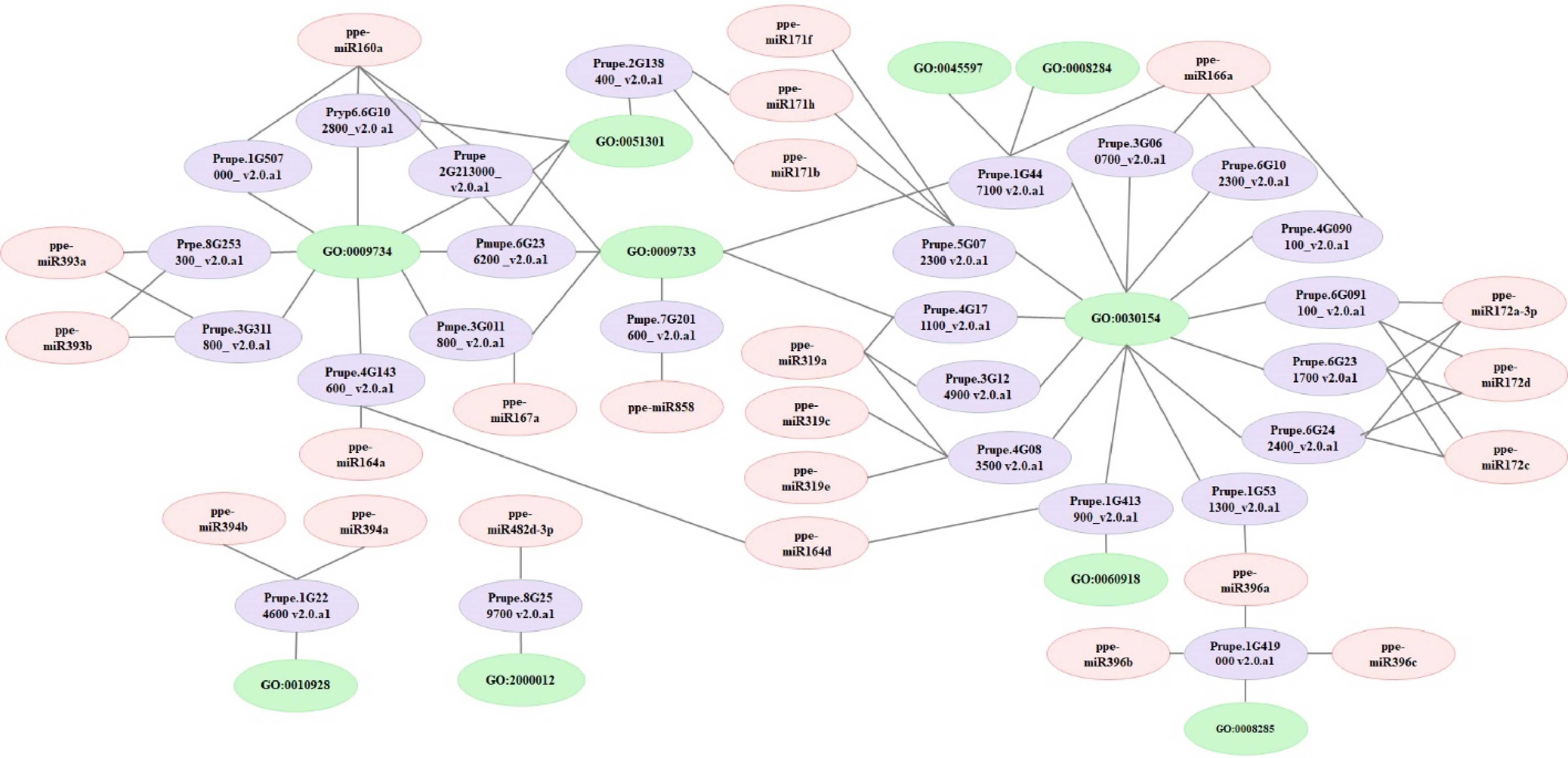
Figure 5.
Network analysis of miRNAs, targets, and GO terms involved in auxin signalling and fruit enlargement. Network analyses were performed using the Cytoscape network platform.
-
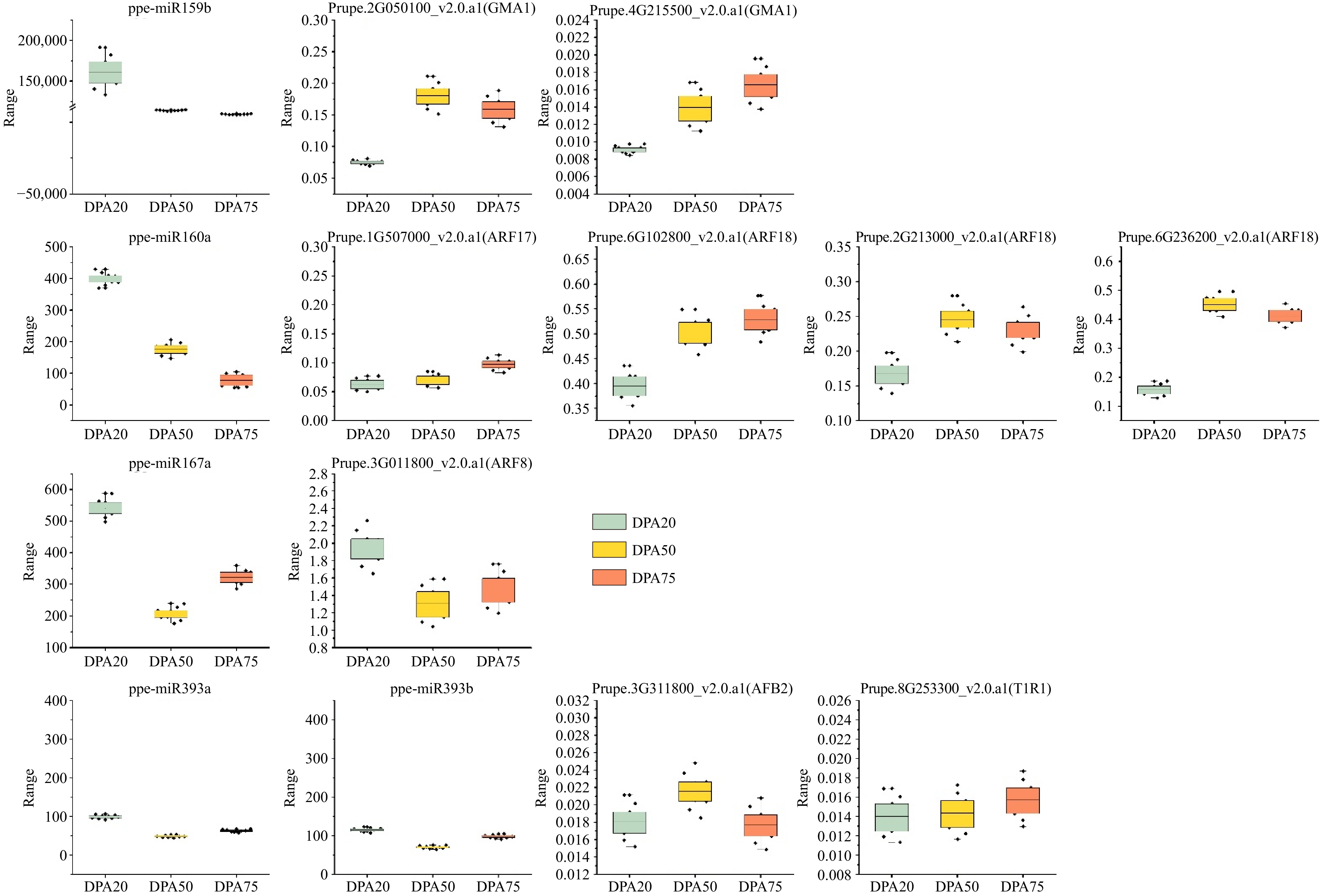
Figure 6.
Expression patterns of the four miRNAs identified and their target genes during fruit development and ripening; U6 and RPII were used as reference genes for miRNA and target genes, respectively.
-
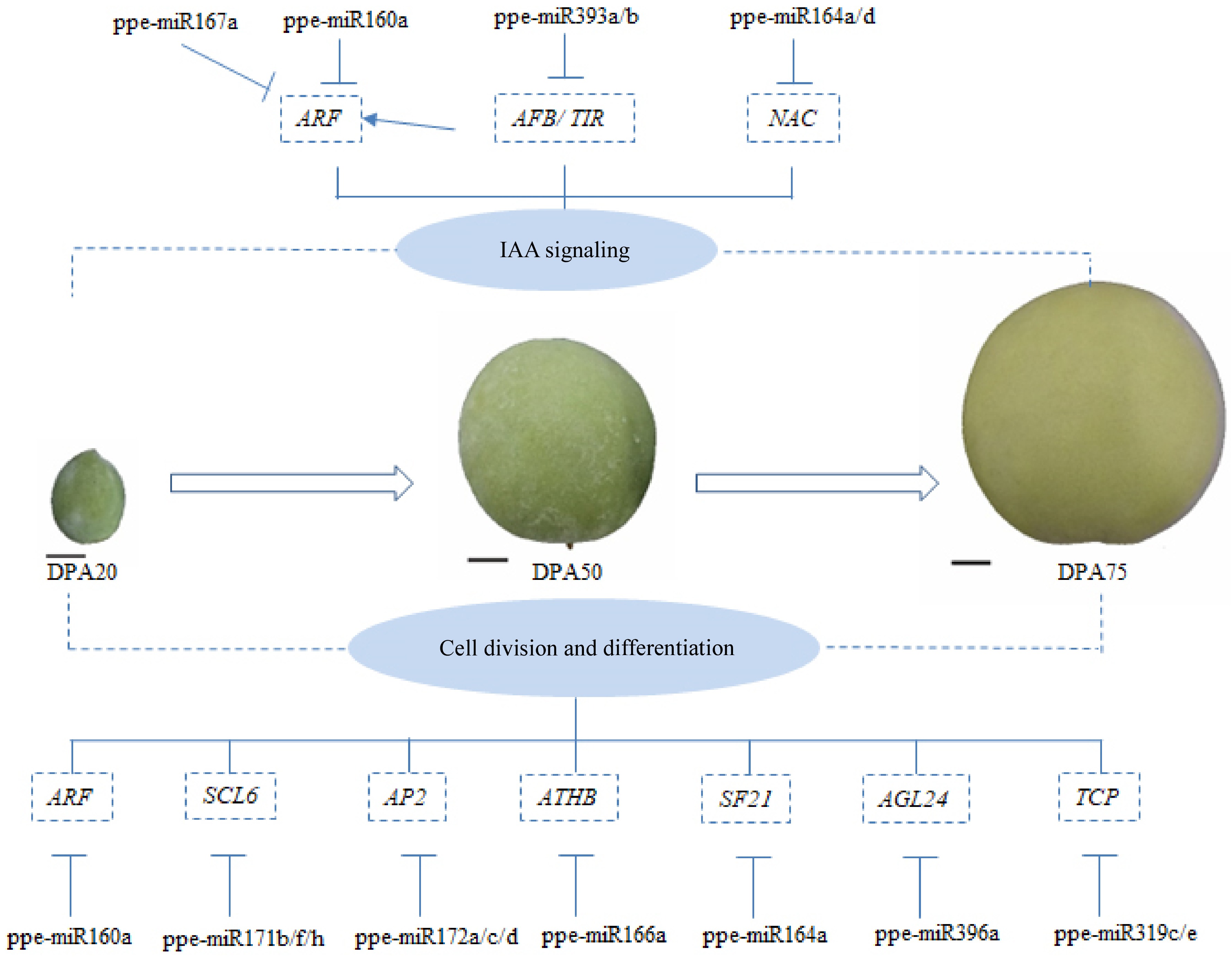
Figure 7.
A schematic model of the proposed roles of miRNAs in the early development of peaches; T bars and arrows refer to negative and positive effects on downstream effectors or biological process, respectively; scale bar = 1 cm
-
miRNA ID miRNA sequences (5'-3') LT1 TPM2 DPA20 DPA50 DPA75 ppe-miR1511-5p CGUGGUAUCAGAGUCAUGUUA 21 154,692.1383 14,578.83618 17,587.11608 ppe-miR1511-3p ACCUGGCUCUGAUACCAUAAC 21 21.90796464 20.71184303 44.79237715 ppe-miR156a UGACAGAAGAAAGAGAGCAC 20 0 4.602631785 26.46822286 ppe-miR156c UGACAGAAGAGAGUGAGCAC 20 0 16.10921125 22.39618858 ppe-miR156f UGACAGAAGAUAGAGAGCAC 20 0 0 10.18008572 ppe-miR159a UUUGGAUUGAAGGGAGCUCUA 21 217,885.6623 131,131.2809 98,590.05813 ppe-miR159b CUUGGAUUGAAGGGAGCUCCA 21 381,669.606 15,059.8112 12,138.73421 ppe-miR160a UGCCUGGCUCCCUGUAUGCCA 21 1,084.44425 333.6908044 177.1334915 ppe-miR162 UCGAUAAACCUCUGCAUCCAG 21 9,792.860194 7,748.53061 4,200.303367 ppe-miR164a UGGAGAAGCAGGGCACGUGCA 21 613.4230099 2,285.206681 804.2267716 ppe-miR164d UGGAGAAGCAGGGCACAUGCU 21 10.95398232 2.301315892 0 ppe-miR166a UCGGACCAGGCUUCAUUCCCC 21 18,358.87437 7,847.487193 3,273.915566 ppe-miR167a UGAAGCUGCCAGCAUGAUCUA 21 1,424.017702 352.1013315 677.9937087 ppe-miR167c UGAAGCUGCCAGCAUGAUCUGA 22 3,768.169918 844.5829325 659.6695544 ppe-miR167d UGAAGCUGCCAGCAUGAUCUUA 22 1,577.373454 342.896068 494.7521658 ppe-miR168 UCGCUUGGUGCAGGUCGGGAA 21 492.9292044 92.0526357 89.5847543 ppe-miR169f UAGCCAAGGAUGACUUGCCUGC 22 0 2.301315892 0 ppe-miR171b UUGAGCCGCGCCAAUAUCACU 21 120.4938055 23.01315892 30.54025715 ppe-miR171d-5p UGUGAUAUUGGUUCGGUUCAUA 22 10.95398232 11.50657946 22.39618858 ppe-miR171d-3p CGAGCCGAAUCAAUAUCACUC 21 10.95398232 4.602631785 16.28813715 ppe-miR171e UUAUUGAACCGGACCAAUAUC 21 10.95398232 2.301315892 0 ppe-miR171f UGAUUGAGCCGUGCCAAUAUC 21 295.7575226 133.4763218 67.18856573 ppe-miR171h UUGAGCCGCGUCAAUAUCUCC 21 635.3309746 211.7210621 24.43220572 ppe-miR172a-3p AGAAUCUUGAUGAUGCUGCAU 21 120.4938055 20.71184303 12.21610286 ppe-miR172c GGAAUCUUGAUGAUGCUGCAU 21 32.86194696 4.602631785 16.28813715 ppe-miR172d GGAAUCUUGAUGAUGCUGCAG 21 65.72389392 2.301315892 4.072034287 ppe-miR2111a UAAUCUGCAUCCUGAGGUUUA 21 0 0 2.036017143 ppe-miR319a UUGGACUGAAGGGAGCUCCC 20 164.3097348 29.9171066 171.02544 ppe-miR319b UAGCUGCCGAGUCAUUCAUCCA 22 0 0 2.036017143 ppe-miR319c UUUGGACUGAAGGGAGCUCC 20 54.7699116 29.9171066 24.43220572 ppe-miR319d CUUGGACUGAAGGGAGCUCCC 21 251.9415934 20.71184303 16.28813715 ppe-miR319e CUUGGACUGAAGGGAGCUCCU 21 328.6194696 36.82105428 81.44068573 ppe-miR319f UUUGGACUGAAGGGAGCUCUC 21 109.5398232 126.5723741 77.36865144 ppe-miR3627-5p UCGCAGGAGAGAUGGCACUGUC 22 43.81592928 6.903947677 2.036017143 ppe-miR390 AAGCUCAGGAGGGAUAGCGCC 21 153.3557525 2.301315892 0 ppe-miR393a CAUCCAAAGGGAUCGCAUUGA 21 262.8955757 87.45000391 136.4131486 ppe-miR393b UCCAAAGGGAUCGCAUUGAUC 21 306.711505 128.87369 215.8178172 ppe-miR394a UUGGCAUUCUGUCCACCUCC 20 3,308.102661 3,516.410684 3,249.483361 ppe-miR394b UUGGCAUUCUGUCCACCUCC 20 54.7699116 39.12237017 44.79237715 ppe-miR395a-3p CUGAAGUGUUUGGGGGGACCC 21 898.2265503 2,908.863288 1,667.49804 ppe-miR395c CUGAAGUGUUUGGGGGAACUC 21 65.72389392 174.9000078 215.8178172 ppe-miR396a UUCCACAGCUUUCUUGAACGU 21 6,364.263728 59,399.2645 71,602.65089 ppe-miR396b UUCCACAGCUUUCUUGAACUU 21 61,791.41427 484,650.223 540,299.9053 ppe-miR396c UUCCACAGCUUUCUUGAACUG 21 12,421.81595 118,018.3829 109,040.9341 ppe-miR397 UCAUUGAGUGCAGCGUUGAUG 21 43.81592928 85.14868802 46.8283943 ppe-miR398a-5p GGAGCGACCUGGGAUCACAUG 21 6,484.757534 11,021.00181 2,640.714235 ppe-miR398a-3p UGUGUUCUCAGGUCGCCCCUG 21 109.5398232 71.34079267 59.04449715 ppe-miR398b CGUGUUCUCAGGUCGCCCCUG 21 1,654.05133 35,502.40027 37,275.40186 ppe-miR398c UGUGUUCUCAGGUCACCCCUU 21 32.86194696 55.23158142 22.39618858 ppe-miR399a CGCCAAAGGAGAGUUGCCCUU 21 0 34.51973839 114.01696 ppe-miR399b UCUGCCAAAGGAGAAUUGCCC 21 32.86194696 27.61579071 73.29661716 ppe-miR399c UGCCAAAGAAGAGUUGCCCUA 21 0 11.50657946 8.144068573 ppe-miR403 UUAGAUUCACGCACAAACUCG 21 230.0336287 191.0092191 425.5275829 ppe-miR477-5p ACUCUCCCUCAAAGGCUUCUAG 22 0 6.903947677 8.144068573 ppe-miR477-3p CGAAGCCUUUGGGGAGAGUAA 21 87.63185856 561.5210778 486.6080972 ppe-miR477a-3p GUUGGGGGCUCUUUUGGGACG 21 65.72389392 987.2645179 787.9386344 ppe-miR477a-5p UCCCUCAAGGGCUCCCAAUAUU 22 0 64.43684499 103.8368743 ppe-miR482a-3p UUUCCGAAACCUCCCAUUCCAA 22 3,833.893812 697.2987154 238.2140058 ppe-miR482a-5p GGGUGAGAGGUUGCCGGAAAGA 22 449.1132751 98.95658337 42.75636001 ppe-miR482b-5p GGAAUGGGAGGAUUGGGAAAA 21 12,750.43542 3,960.564651 2,972.585029 ppe-miR482b-3p CUUCCCAAACCUCCCAUUCCUA 22 32.86194696 4.602631785 10.18008572 ppe-miR482c-5p GGAAUGGGCUGUUUGGGAUG 20 755.8247801 409.6342289 191.3856115 ppe-miR482c-3p UUCCCAAGCCCGCCCAUUCCAA 22 3,428.596466 554.6171301 555.8326801 ppe-miR482e UUGCCUAUUCCUCCCAUGCCAA 22 942.0424795 849.1855643 572.1208173 ppe-miR482f UCUUUCCUACUCCACCCAUUCC 22 8,839.863732 10,813.88338 16,467.30665 ppe-miR5225-3p UCAUCUCUCCUCGACUGAA 19 21.90796464 0 0 ppe-miR5225-5p UCUGUCGUAGGAGAGAUGGCGC 22 32.86194696 0 0 ppe-miR530 UCUGCAUUUGCACCUGCACCU 21 87.63185856 23.01315892 30.54025715 ppe-miR535a UGACAACGAGAGAGAGCACGC 21 76.67787624 62.1355291 12.21610286 ppe-miR535b UGACGACGAGAGAGAGCACGC 21 197.1716818 186.4065873 77.36865144 ppe-miR6257 UCUUAACUGUUGGAUUAGGCU 21 10.95398232 11.50657946 2.036017143 ppe-miR6258 UUCCAGCUGUAAAGAUCAAGA 21 10.95398232 6.903947677 12.21610286 ppe-miR6260 UGGAGUGAGAGAAUGGGAGGU 21 0 0 6.10805143 ppe-miR6261 AAGUGAUUAUAUGGAGAAGCAC 22 0 0 2.036017143 ppe-miR6263 AAGUGGACAAAAGGGGAGUGG 21 0 0 2.036017143 ppe-miR6264 AUGCCUAUGGACACGUGUCAA 21 10.95398232 4.602631785 6.10805143 ppe-miR6265 UUGAACUUUGACCCGAUUCGCAU 23 21.90796464 4.602631785 2.036017143 ppe-miR6266a UAAAUGCAGGGGCAAAAUGAU 21 0 2.301315892 0 ppe-miR6267a UAGAGAGGUGGUACAAUUGUG 21 10.95398232 0 0 ppe-miR6267c-5p AUUGCUGAUCACCUCUCUAAU 21 21.90796464 59.8342132 185.27756 ppe-miR6267c-3p UAGAGAGAUGGUCAGCAAUGU 21 21.90796464 27.61579071 105.8728914 ppe-miR6270 UUCUGGUAUUGGAAUUUCAUU 21 142.4017702 18.41052714 40.72034287 ppe-miR6271 UCAAGAUUGAGAGAUAUAAUG 21 0 0 8.144068573 ppe-miR6274b-5p AUUUCGACUAAUAACACAAUG 21 0 2.301315892 2.036017143 ppe-miR6274b-3p UUGUGUUAUUGGCCGAAAAUAG 22 208.1256641 18.41052714 46.8283943 ppe-miR6277 UGUGUGUGGAAAGAGCGAGAC 21 21.90796464 9.20526357 12.21610286 ppe-miR6280 UUGGCAGUAAGAUUUUUGGUG 21 10.95398232 0 0 ppe-miR6281 GUUAGAGAUAGAGAGAGUGAG 21 76.67787624 52.93026553 69.22458287 ppe-miR6282 GUUGAUCGAUGUGGGAUGUUACA 23 10.95398232 0 0 ppe-miR6283 CAAAAGGGGAGUGGGAAAAUC 21 21.90796464 0 0 ppe-miR6284 UUUGGACCAUGGAUGAAGAUU 21 98.58584088 29.9171066 20.36017143 ppe-miR6285 UAGUGAAGUUUGAAUUAGGGCU 22 0 2.301315892 0 ppe-miR6287 CAAGAAGUGGAAGUUUUGGGC 21 0 0 2.036017143 ppe-miR6288a GAAAAUGACAAGUGGCUAGUU 21 0 2.301315892 2.036017143 ppe-miR6288b-3p UCAAUUAGAAAAUGAUAAGUG 21 0 0 4.072034287 ppe-miR6290 UGAAUGAGUUCAGAGAUCGUGUA 23 0 0 2.036017143 ppe-miR6292 UAUCUUUUAAUCGUUAGAUCA 21 0 0 2.036017143 ppe-miR6293 UAAGAGGCUGAUGACUAAAAC 21 65.72389392 46.02631785 61.0805143 ppe-miR6294 UGGUGUAGGCUAAUCACAAUC 21 32.86194696 20.71184303 42.75636001 ppe-miR6295 GAGGACAGAAGAUGAUUCAGC 21 32.86194696 13.80789535 20.36017143 ppe-miR6297a AAUAAUUUUUCGUCGCGCAAAAU 23 32.86194696 20.71184303 8.144068573 ppe-miR6297b GAUGUAUUGUCGUCGCGCAAAGU 23 0 0 2.036017143 ppe-miR7122a-5p UUAUACAAUGAAAUCACGGCCG 22 251.9415934 69.03947677 83.47670287 ppe-miR7122a-3p GCCGUGUUUCUUUGUAUAAAG 21 1,237.800002 1,456.73296 2,158.178172 ppe-miR7122b-3p CCGUGUUUCCUUGUAUAAAG 20 0 2.301315892 8.144068573 ppe-miR7122b-5p UUAUACAAUGAAAUCACGGUCG 22 32.86194696 46.02631785 140.4851829 ppe-miR7125-3p CGAACUUAUUGCAACUAGCUU 21 0 27.61579071 8.144068573 ppe-miR8122-5p UUCCACAGAUCUUUCCUCAUU 21 5,466.037178 1,233.505318 1,284.726817 ppe-miR8122-3p UGAAGGAAGAUUUGUGGAAAG 21 887.2725679 667.3816088 637.2733658 ppe-miR8124-3p UGGCACCAAUGAUACCAAGUUU 22 54.7699116 11.50657946 4.072034287 ppe-miR8124-5p ACUUGGUAUCUUGGUGCCGGU 21 0 2.301315892 2.036017143 ppe-miR8125 CAGGAAAGAAUGUGAUGAGUA 21 65.72389392 27.61579071 46.8283943 ppe-miR8126-5p UCUGAGUCAGAUUACUGAAUA 21 10.95398232 4.602631785 8.144068573 ppe-miR8126-3p UUCAGUAUUUUGACUCAGAA 20 21.90796464 23.01315892 42.75636001 ppe-miR8127-3p UUCAAAGGGUACAUCCACAGU 21 10.95398232 0 0 ppe-miR8128-5p AUUAGACCUCUCCCGACGAAA 21 21.90796464 43.72500196 63.11653144 ppe-miR8128-3p UCGUGGGGAGAGAUCUAAUCG 21 0 0 4.072034287 ppe-miR8130-5p GGGUUCCUUGUUGGAAGGACU 21 0 2.301315892 0 ppe-miR8131-5p AUUUCAGCUAAGUUGAGUUGU 21 10.95398232 0 2.036017143 ppe-miR8132 UCCAACGAUGGGUGACCACAA 21 10.95398232 2.301315892 10.18008572 ppe-miR8133-5p UCCUGUGCGAACGUCCAGAAG 21 87.63185856 46.02631785 28.50424001 ppe-miR8133-3p UAACUUCCGAACGUCCGCAUA 21 32.86194696 16.10921125 12.21610286 ppe-miR827 UUAGAUGACCAUCAACAAACA 21 1,084.44425 409.6342289 309.4746058 ppe-miR858 CUCGUUGUCUGUUCGACCUUG 21 10.95398232 4.602631785 2.036017143 1 Length of miRNA; 2 the expression of transcripts per million. Table 1.
Characteristics of know miRNAs identified from DPA20, DAP50, and DPA75 libraries in peach fruit.
-
GO_Id GO_term miRNA DPA20_TPM DPA50_TPM DPA75_TPM Transcript_ID Symbol Annoation DPA20_TPM DPA50_TPM DPA75_TPM GO:0009734 Auxin-activated signaling pathway ppe-miR160a 1,084.44425 333.6908044 177.1334915 Prupe.1G507000_v2.0.a1 ARF17 Auxin response factor 3 2,936.993249 2,693.359061 5,571.987995 Prupe.2G213000_v2.0.a1 ARF18 Auxin response factor 3 4,104.259796 9,286.058852 11,365.58915 Prupe.6G102800_v2.0.a1 ARF18 Hypothetical protein 78,884.62636 112,477.8903 81,142.07518 Prupe.6G236200_v2.0.a1 ARF18 Auxin response factor 3 10,806.629 33,566.48979 26,055.37568 ppe-miR164a 613.4230099 2,285.206681 804.2267716 Prupe.4G143600_v2.0.a1 NAC021 NAC domain-containing protein 89-like 489.4988748 8,562.46985 5,160.4207 ppe-miR164d 10.95398232 2.301315892 0 Prupe.4G143600_v2.0.a1 NAC021 NAC domain-containing protein 89-like 489.4988748 8,562.46985 5,160.4207 ppe-miR167a 1,424.017702 352.1013315 677.9937087 Prupe.3G011800_v2.0.a1 ARF8 Auxin response factor 19-like 1,845.03422 4,502.331565 12,663.60908 ppe-miR393a 262.8955757 87.45000391 136.4131486 Prupe.3G311800_v2.0.a1 AFB2 Protein TRANSPORT INHIBITOR RESPONSE 1-like 0 120.5981669 0 Prupe.8G253300_v2.0.a1 TIR1 Hypothetical protein 37.6537596 120.5981669 221.6131589 ppe-miR393b 306.711505 128.87369 215.8178172 Prupe.3G311800_v2.0.a1 AFB2 Protein TRANSPORT INHIBITOR RESPONSE 1-like 4,367.836113 9,848.850297 19,406.98092 Prupe.8G253300_v2.0.a1 TIR1 Hypothetical protein 6,137.562814 32,923.29957 57,841.03448 GO:0010928 regulation of auxin mediated signaling pathway ppe-miR394b 54.7699116 39.12237017 44.79237715 Prupe.1G224600_v2.0.a1 FBX6 − 978.9977495 4,140.537064 7,408.211312 ppe-miR394a 3,308.102661 3,516.410684 3,249.483361 Prupe.1G224600_v2.0.a1 FBX6 − 978.9977495 4,140.537064 7,408.211312 GO:0009733 response to auxin ppe-miR167a 1424.017702 352.1013315 677.9937087 Prupe.3G011800_v2.0.a1 ARF8 Auxin response factor 19-like 1,845.03422 4,502.331565 12,663.60908 ppe-miR160a 1,084.44425 333.6908044 177.1334915 Prupe.2G213000_v2.0.a1 ARF18 Auxin response factor 3 4,104.259796 9,286.058852 11,365.58915 Prupe.6G236200_v2.0.a1 ARF18 Auxin response factor 3 10,806.629 33,566.48979 26,055.37568 ppe-miR858 10.95398232 4.602631785 2.036017143 Prupe.7G201600_v2.0.a1 C1 Transcription factor MYB75-like 0 40.19938897 886.4526356 ppe-miR166a 18,358.87437 7,847.487193 3,273.915566 Prupe.1G447100_v2.0.a1 ATHB-8 − 414.1913556 2,170.767004 2,421.915237 ppe-miR319a 164.3097348 29.9171066 171.02544 Prupe.4G171100_v2.0.a1 PCF5 − 112.9612788 0 94.9770681 GO:0060918 Auxin transport ppe-miR164d 10.95398232 2.301315892 0 Prupe.1G413900_v2.0.a1 SF21 − 37.6537596 0 0 GO:2000012 Regulation of auxin polar transport ppe-miR482d-3p 0 0 0 Prupe.8G259700_v2.0.a1 OsI_027940 Uncharacterized protein OsI_027940-like 37.6537596 120.5981669 31.6590227 GO:0030154 Cell differentiation ppe-miR319c 54.7699116 29.9171066 24.43220572 Prupe.4G083500_v2.0.a1 TCP2 − 0 0 63.3180454 ppe-miR319e 328.6194696 36.82105428 81.44068573 Prupe.4G083500_v2.0.a1 TCP2 − 0 0 63.3180454 ppe-miR164d 10.95398232 2.301315892 0 Prupe.1G413900_v2.0.a1 SF21 − 37.6537596 0 0 ppe-miR166a 18,358.87437 7,847.487193 3,273.915566 Prupe.1G447100_v2.0.a1 ATHB-8 − 414.1913556 2,170.767004 2,421.915237 Prupe.3G060700_v2.0.a1 ATHB-15 − 414.1913556 2,170.767004 2,421.915237 Prupe.4G090100_v2.0.a1 ATHB-14 − 941.3439899 924.5859463 569.8624086 Prupe.6G102300_v2.0.a1 REV − 715.4214324 1,165.78228 1,677.928203 ppe-miR171b 120.4938055 23.01315892 30.54025715 Prupe.5G072300_v2.0.a1 SCL6 Hypothetical protein 715.4214324 442.1932787 348.2492497 ppe-miR171f 295.7575226 133.4763218 67.18856573 Prupe.5G072300_v2.0.a1 SCL6 Hypothetical protein 0 241.1963338 1,171.38384 ppe-miR171h 635.3309746 211.7210621 24.43220572 Prupe.5G072300_v2.0.a1 SCL6 Hypothetical protein 715.4214324 442.1932787 348.2492497 ppe-miR172a-3p 120.4938055 20.71184303 12.21610286 Prupe.6G091100_v2.0.a1 RAP2-7 − 37.6537596 200.9969448 284.9312043 Prupe.6G231700_v2.0.a1 AP2 Uncharacterized LOC106378603 376.537596 1,045.184113 3,609.128588 Prupe.6G242400_v2.0.a1 AP2 − 0 0 94.9770681 ppe-miR172c 32.86194696 4.602631785 16.28813715 Prupe.6G091100_v2.0.a1 RAP2-7 − 37.6537596 200.9969448 284.9312043 Prupe.6G231700_v2.0.a1 AP2 Uncharacterized LOC106378603 376.537596 1,045.184113 3,609.128588 Prupe.6G242400_v2.0.a1 AP2 − 0 0 94.9770681 ppe-miR172d 65.72389392 2.301315892 4.072034287 Prupe.6G091100_v2.0.a1 RAP2-7 − 37.6537596 200.9969448 284.9312043 Prupe.6G231700_v2.0.a1 AP2 Uncharacterized LOC106378603 376.537596 1,045.184113 3,609.128588 Prupe.6G242400_v2.0.a1 AP2 − 0 0 94.9770681 ppe-miR319a 164.3097348 29.9171066 171.02544 Prupe.3G124900_v2.0.a1 TCP4 − 37.6537596 0 63.3180454 Prupe.4G083500_v2.0.a1 TCP2 − 75.3075192 80.39877794 664.8394767 Prupe.4G171100_v2.0.a1 PCF5 − 112.9612788 0 94.9770681 ppe-miR396a 6364.263728 59399.2645 71602.65089 Prupe.1G531300_v2.0.a1 AGL24 D-3-phosphoglycerate dehydrogenase 0 93.79857426 164.626918 GO:0045597 Positive regulation of cell differentiation ppe-miR166a 18,358.87437 7,847.487193 3,273.915566 Prupe.1G447100_v2.0.a1 ATHB-8 − 414.1913556 2,170.767004 2,421.915237 GO:0051301 Cell division ppe-miR160a 1,084.44425 333.6908044 177.1334915 Prupe.2G213000_v2.0.a1 ARF18 Auxin response factor 3 4,104.259796 9,286.058852 11,365.58915 Prupe.6G102800_v2.0.a1 ARF18 Hypothetical protein 78,884.62636 112,477.8903 81,142.07518 Prupe.6G236200_v2.0.a1 ARF18 Auxin response factor 3 10,806.629 33,566.48979 26,055.37568 ppe-miR171b 120.4938055 23.01315892 30.54025715 Prupe.2G138400_v2.0.a1 SCL6 DELLA protein GAI 1 37.6537596 0 0 ppe-miR171h 635.3309746 211.7210621 24.43220572 Prupe.2G138400_v2.0.a1 SCL6 DELLA protein GAI 1 37.6537596 0 0 GO:0008284 Positive regulation of cell proliferation ppe-miR166a 18,358.87437 7,847.487193 3,273.915566 Prupe.1G447100_v2.0.a1 ATHB-8 − 414.1913556 2,170.767004 2,421.915237 GO:0008285 Negative regulation of cell proliferation ppe-miR396a 6,364.263728 59,399.2645 71,602.65089 Prupe.1G419000_v2.0.a1 GRF6 − 1,242.574067 6,070.107734 4,558.899269 ppe-miR396b 61,791.41427 484,650.223 540,299.9053 Prupe.1G419000_v2.0.a1 GRF6 − 1,242.574067 6,070.107734 4,558.899269 ppe-miR396c 12,421.81595 118,018.3829 109,040.9341 Prupe.1G419000_v2.0.a1 GRF6 − 1,242.574067 6,070.107734 4,558.899269 Table 2.
The miRNA, target genes, and GO terms information used for network analysis.
Figures
(7)
Tables
(2)