-
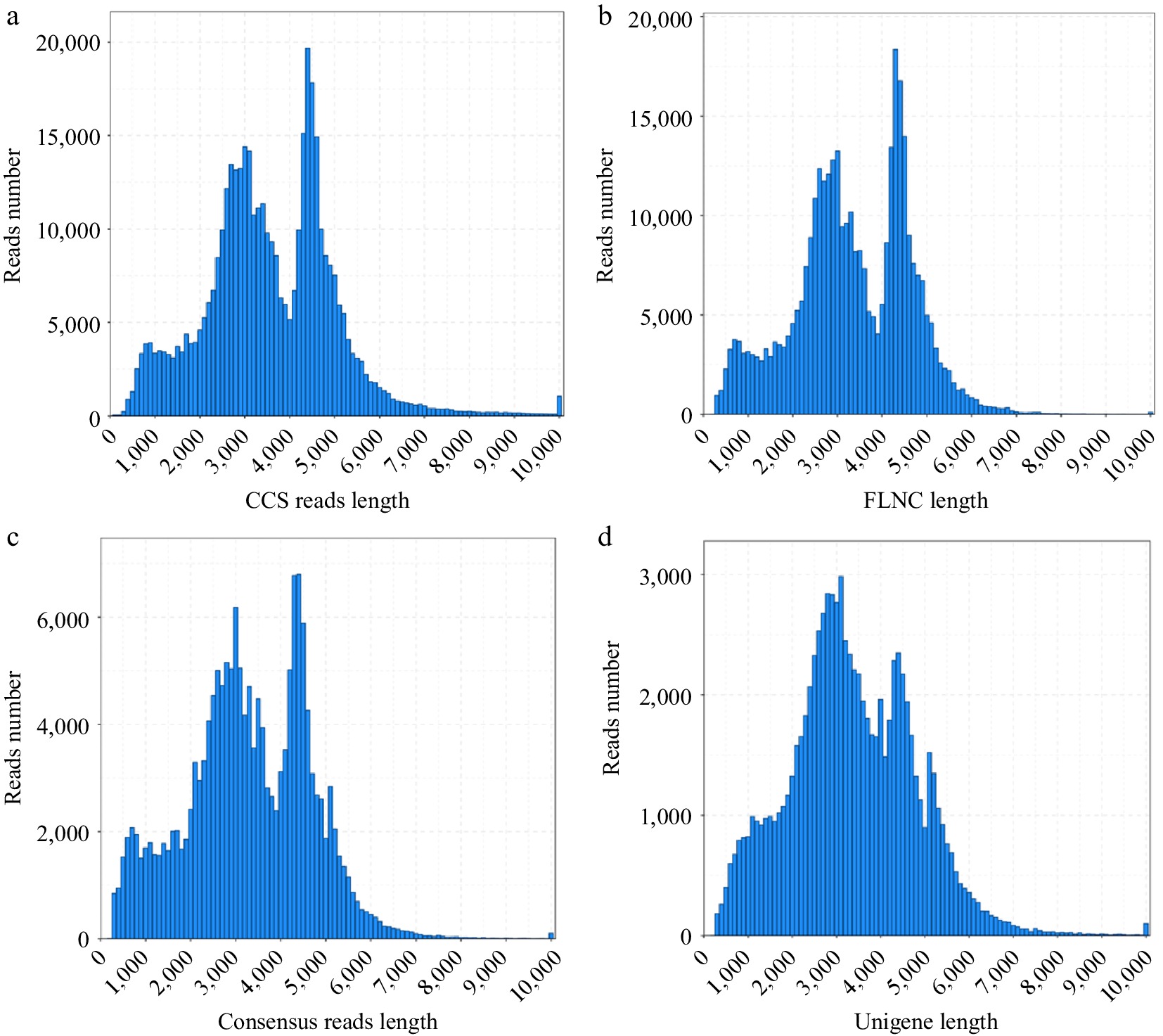
Figure 1.
Length distributions of PacBio SMRT sequences. (a) Number and length distributions of circular consensus sequences. (b) Number and length distributions of full-length non-chimeric sequences. (c) Number and length distributions of consensus isoforms. (d) Number and length distributions of unigenes.
-
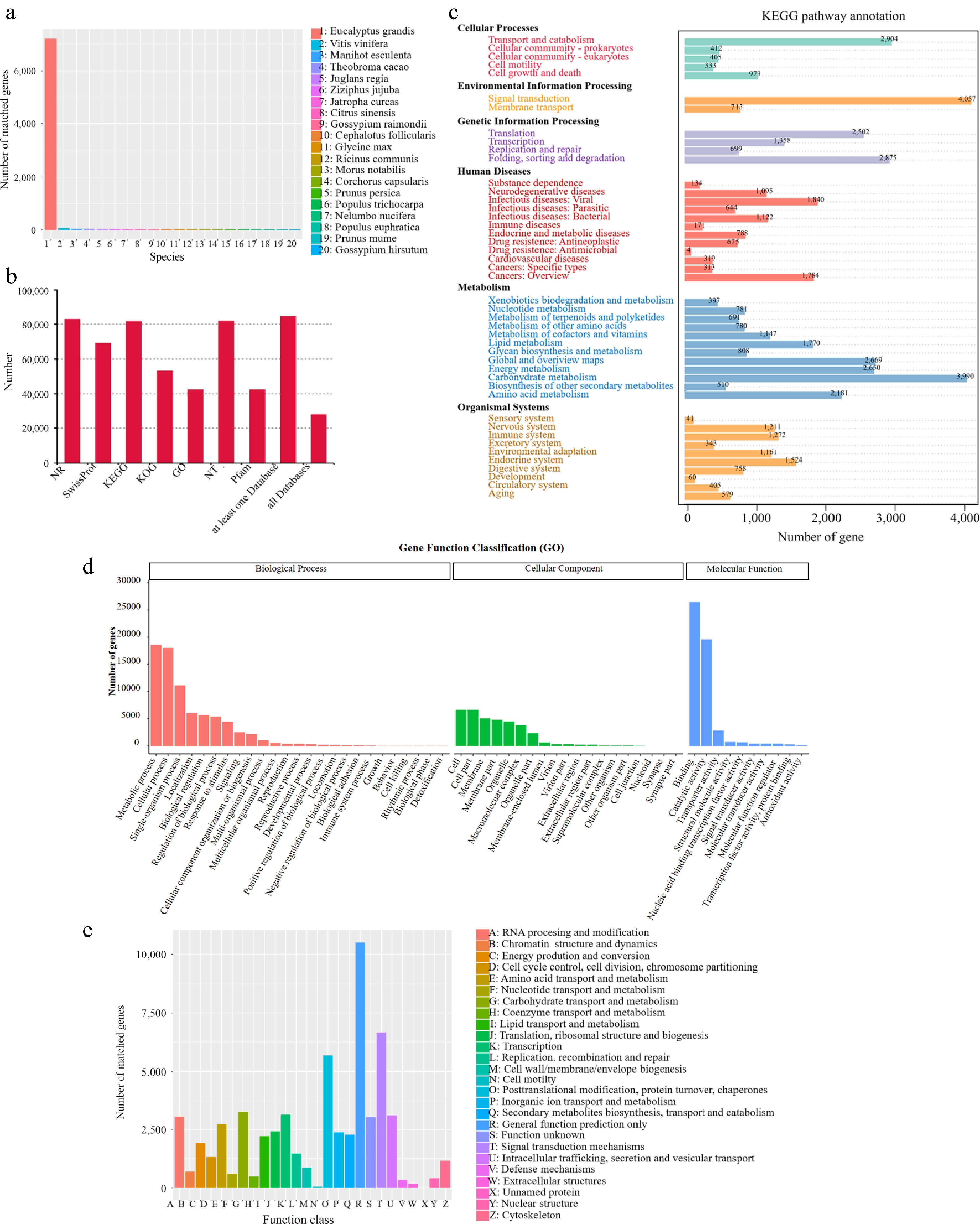
Figure 2.
Annotation of Jaboticaba unigenes. (a) NR homologous species distribution of unigenes. (b) Function annotation of unigenes in all databases. (c) KEGG pathways enriched for unigenes. (d) Distribution of GO terms for all annotated unigenes. (e) Searching of unigenes against the KOG database.
-
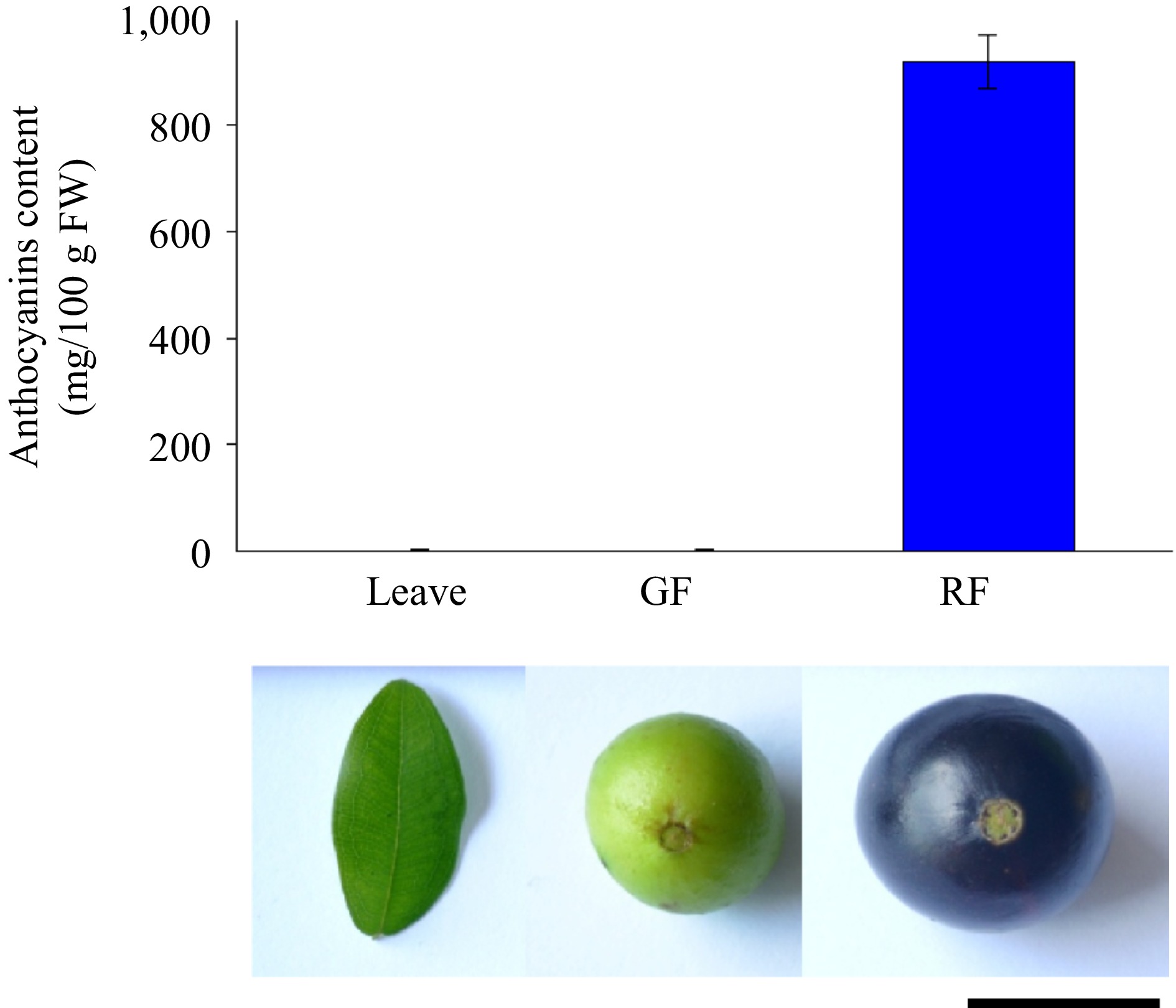
Figure 3.
Anthocyanin content in different tissues of Jaboticaba.
-
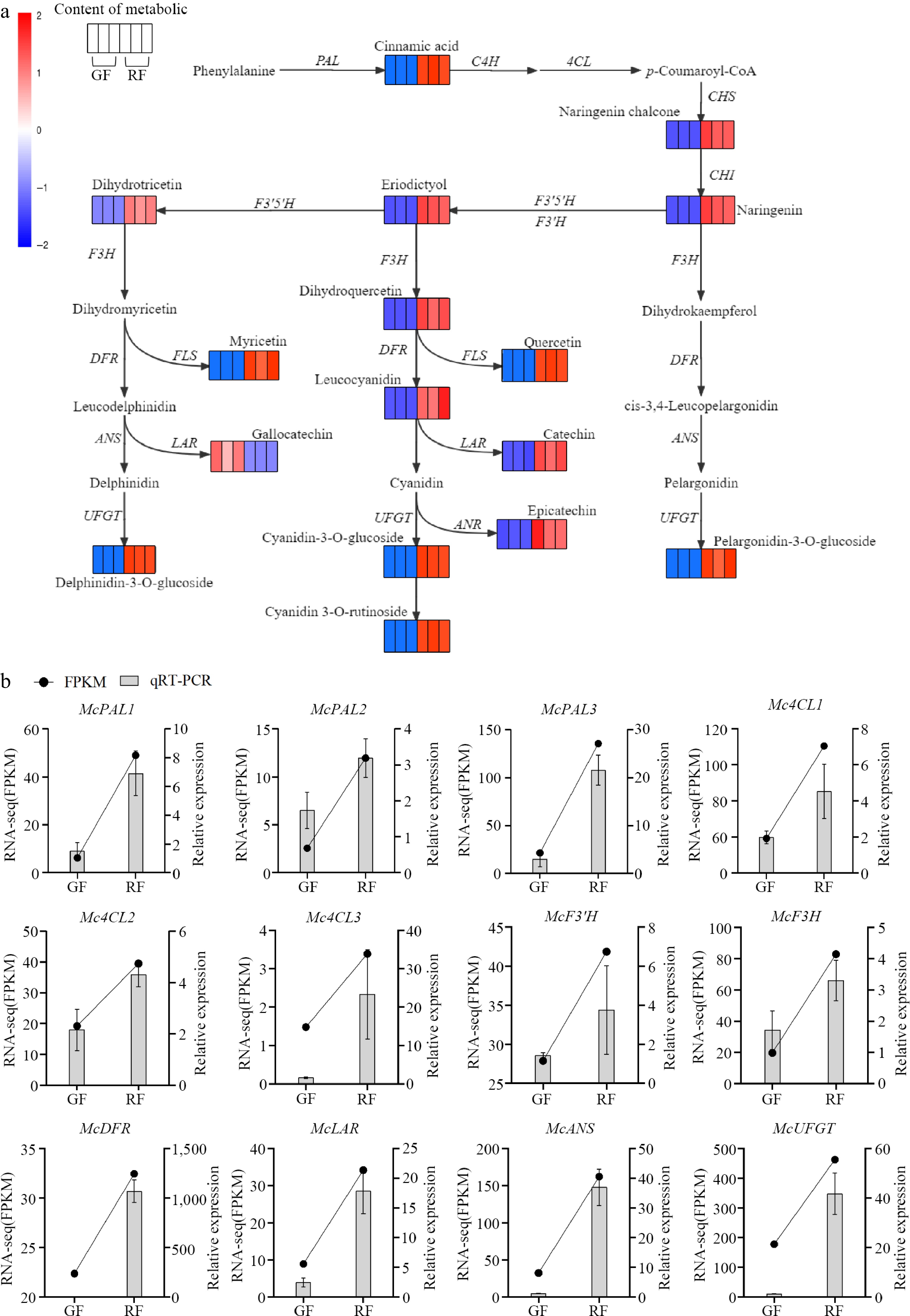
Figure 4.
Anthocyanin biosynthesis pathway in Jaboticaba. (a) Transcript and metabolic profiling of genes and metabolites in the phenylpropanoid, flavonoid and anthocyanin biosynthetic pathways in green fruits (GF) and ripening fruits (RF). Each colored cell represents the log2(FPKM) and log10(Content) value of each pathway gene and metabolite according to the color scale, respectively. PAL, phenylalanine ammonia-lyase; C4H, cinnamic acid 4-hydroxylase; 4CL, 4-coumarate CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3'H, flavanoid 3'-hydroxylase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanidin synthase; UFGT, UDP glucose-flavonoid 3-O-glcosyl-transferase; FLS, flavonol synthesis; LAR, leucocyanidin reductase; ANR, anthocyanin reductase. (b) Expression levels of the color-related genes in green fruits (GF) and ripening fruits (RF) validated by RNA-seq and qRT-PCR.
-
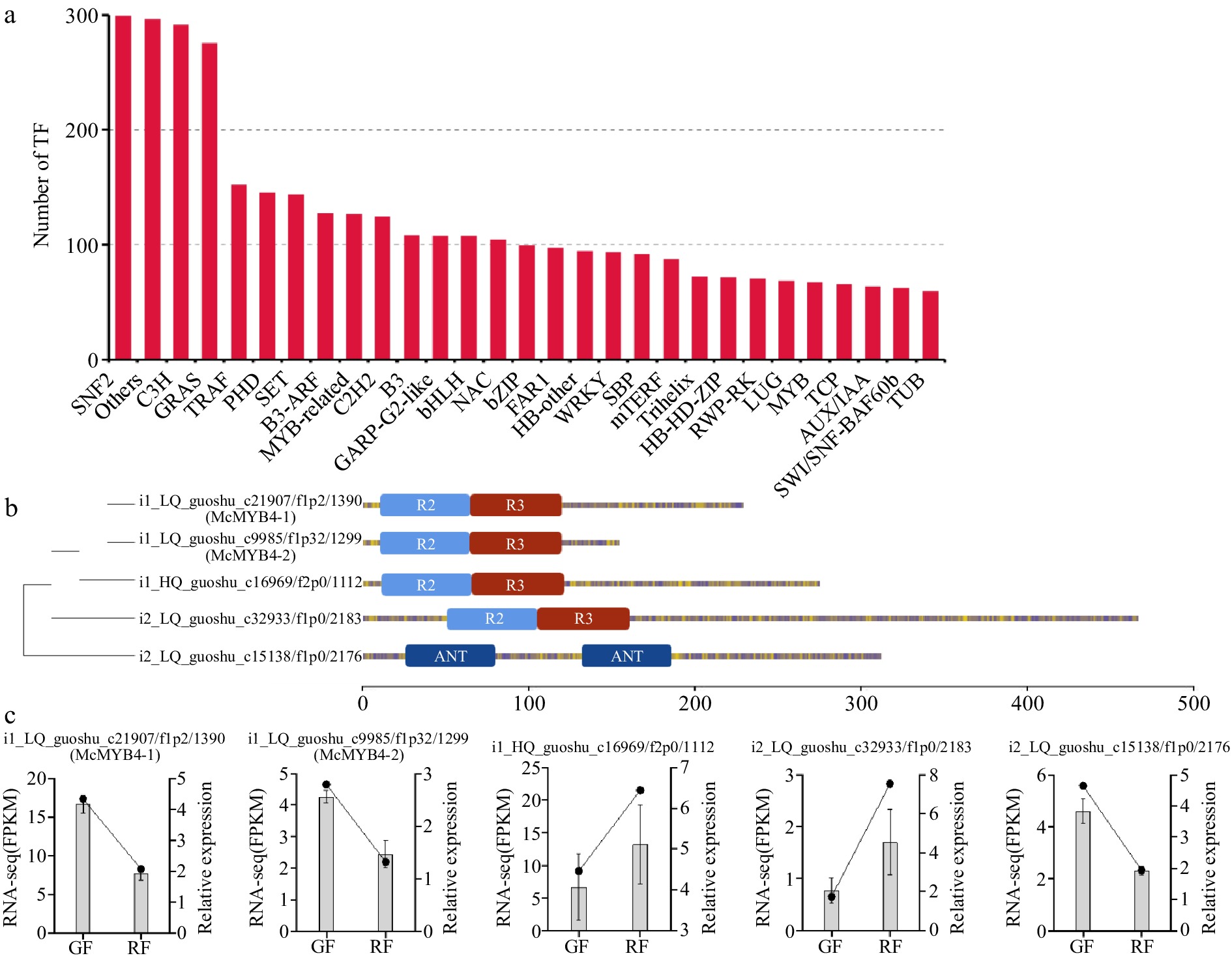
Figure 5.
McMYBs analysis in Jaboticaba. (a) The number of TFs belonging to different TF families. (b) Phylogenetic relationships and gene structure analysis of five McMYBs. (c) Expression levels of the five McMYBs in green fruits (GF), and ripening fruits (RF) validated by RNA-seq and qRT-PCR.
-
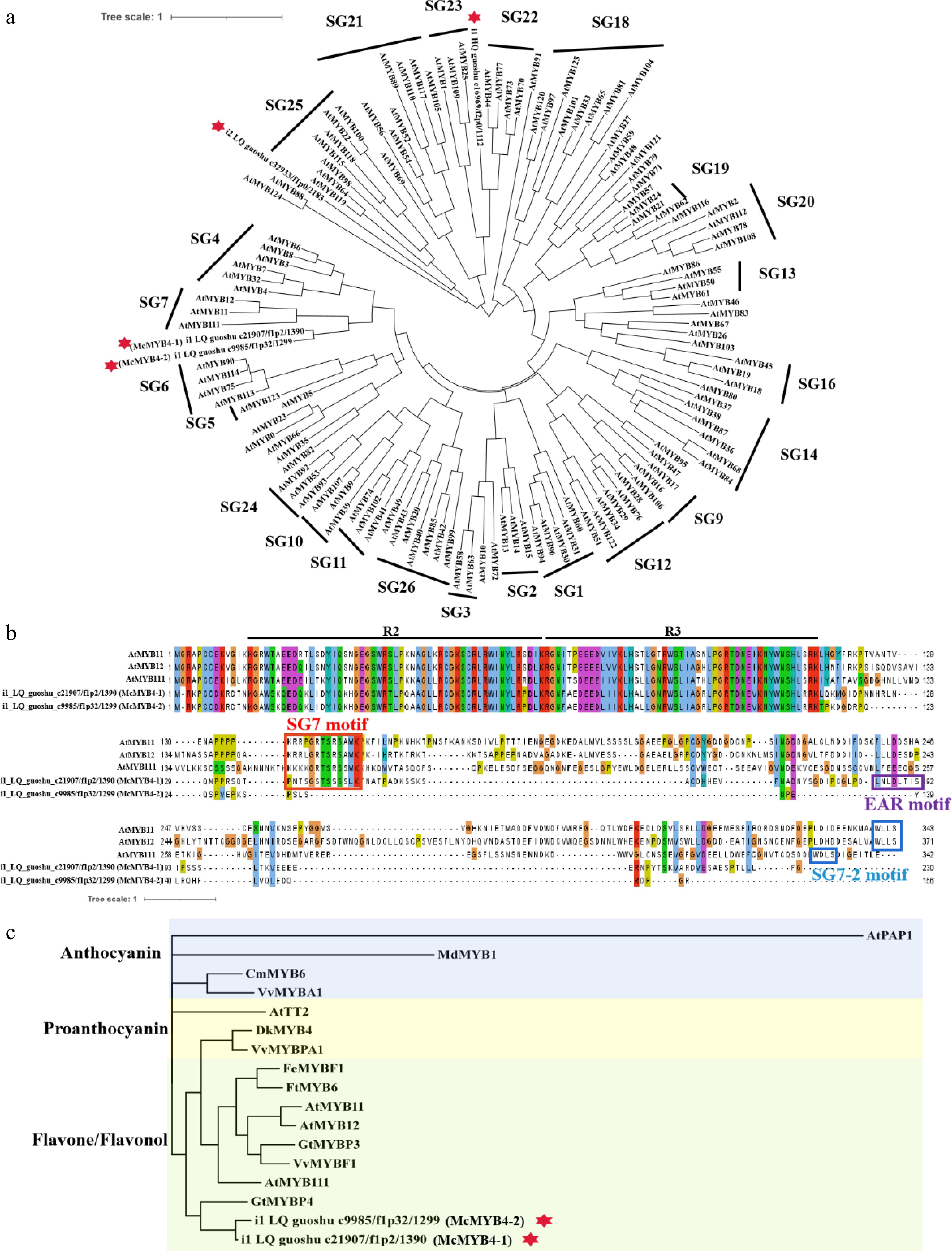
Figure 6.
Subgroup and phylogenetic relationships of McMYBs. (a) Phylogenetic relationships of four McMYBs and R2R3 MYBs from Arabidopsis. A total of 126 protein sequences of the R2R3 MYBs in Arabidopsis were obtained from The Arabidopsis Information Resource (TAIR) database. (b) Multiple alignment of SG7 R2R3-MYB proteins. The R2, R3 MYB domains, SG7, SG7-2 and EAR motif are marked. (c) Phylogenetic relationships of McMYB4-1, McMYB4-2 and R2R3 MYBs from other species. At, Arabidopsis thaliana; Cm, Chrysanthemum morifolium; Dk, Diospyros kaki; Fe, Fagopyrum esculentum; Ft, Fagopyrum tataricum; Gt, Gentiana trifloral; Md, Malus domestica; Vv, Vitis vinifera.
-
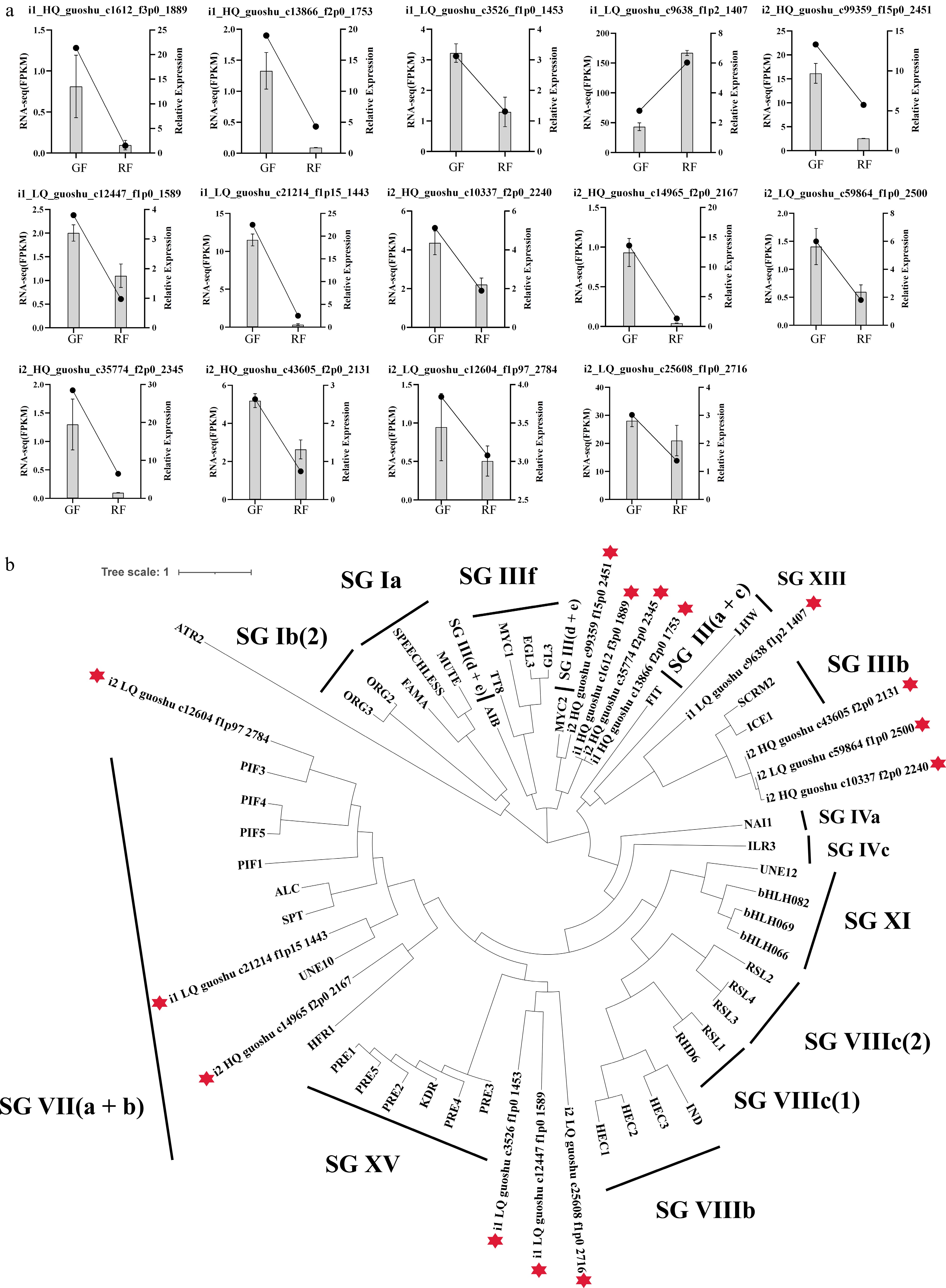
Figure 7.
Expression and phylogenetic analysis of McbHLH transcription factors. (a) Expression levels of the 14 McbHLHs in green fruits (GF), and ripening fruits (RF) validated by RNA-seq and qRT-PCR. (b) Phylogenetic relationships of 14 McbHLHs and bHLHs from Arabidopsis. A total of 45 protein sequences of the bHLHs in Arabidopsis were obtained from the TAIR database.
Figures
(7)
Tables
(0)