-
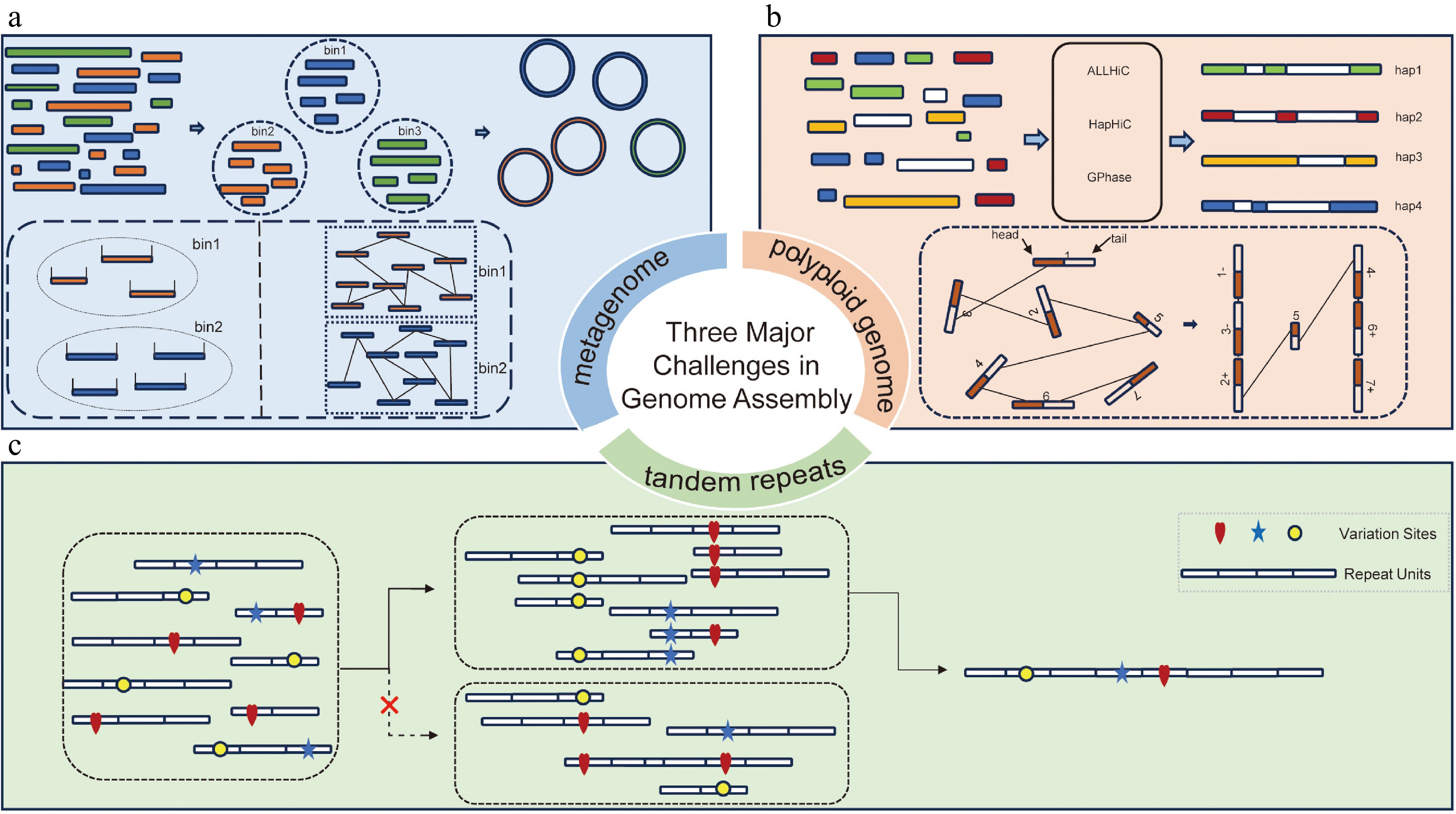
Figure 1.
Schematics of assembling tandem repeats, polyploid genomes, and metagenomes. (a) Schematic of metagenomic binning: assembled contigs are grouped into different bins based on sequencing read coverage (left panel in dashed box) or Hi-C interaction signals (right panel in dashed box). These contigs are then connected to reconstruct genomes as completely as possible. (b) Schematic of haplotype-resolved scaffolding in polyploid genomes: in this step, preliminary contigs are assigned to different chromosomes and their respective haplotypes using Hi-C interaction signals, followed by ordering and orientation (as shown in the dashed box), ultimately achieving haplotype-resolved assembly. (c) Schematics of tandem repeat assembly: reads from tandemly repetitive regions were aligned in a pairwise manner. Sequence variations between repeat units, such as SNPs and small indels, were then used to evaluate the alignment correctness. Only correct alignments were retained and used to assemble the reads into continuous tandem repeat sequences.
-
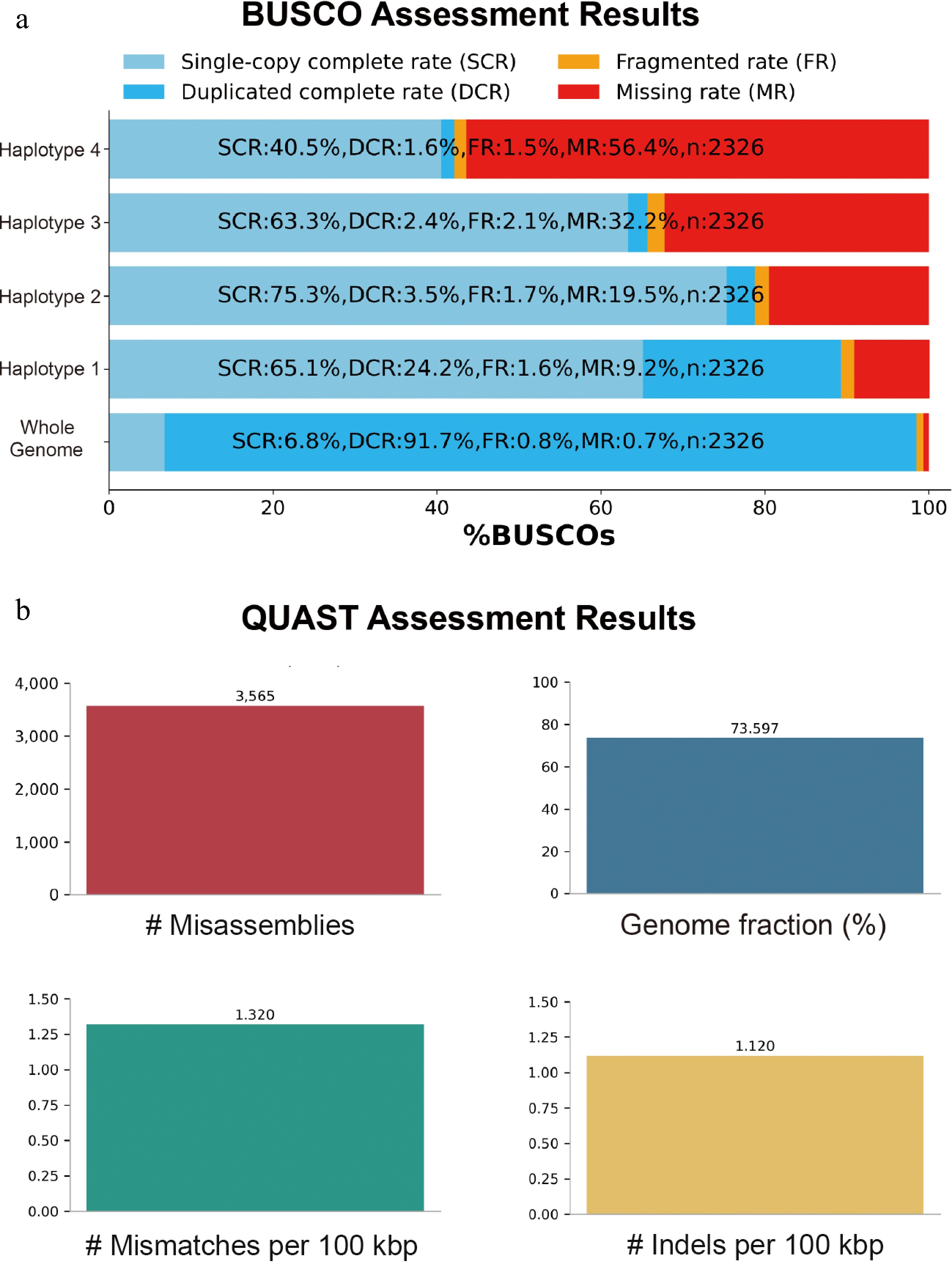
Figure 2.
Genome assembly assessment for the autotetraploid potato cultivar C88. (a) BUSCO completeness scores. (b) QUAST assessment scores.
Figures
(2)
Tables
(0)