-
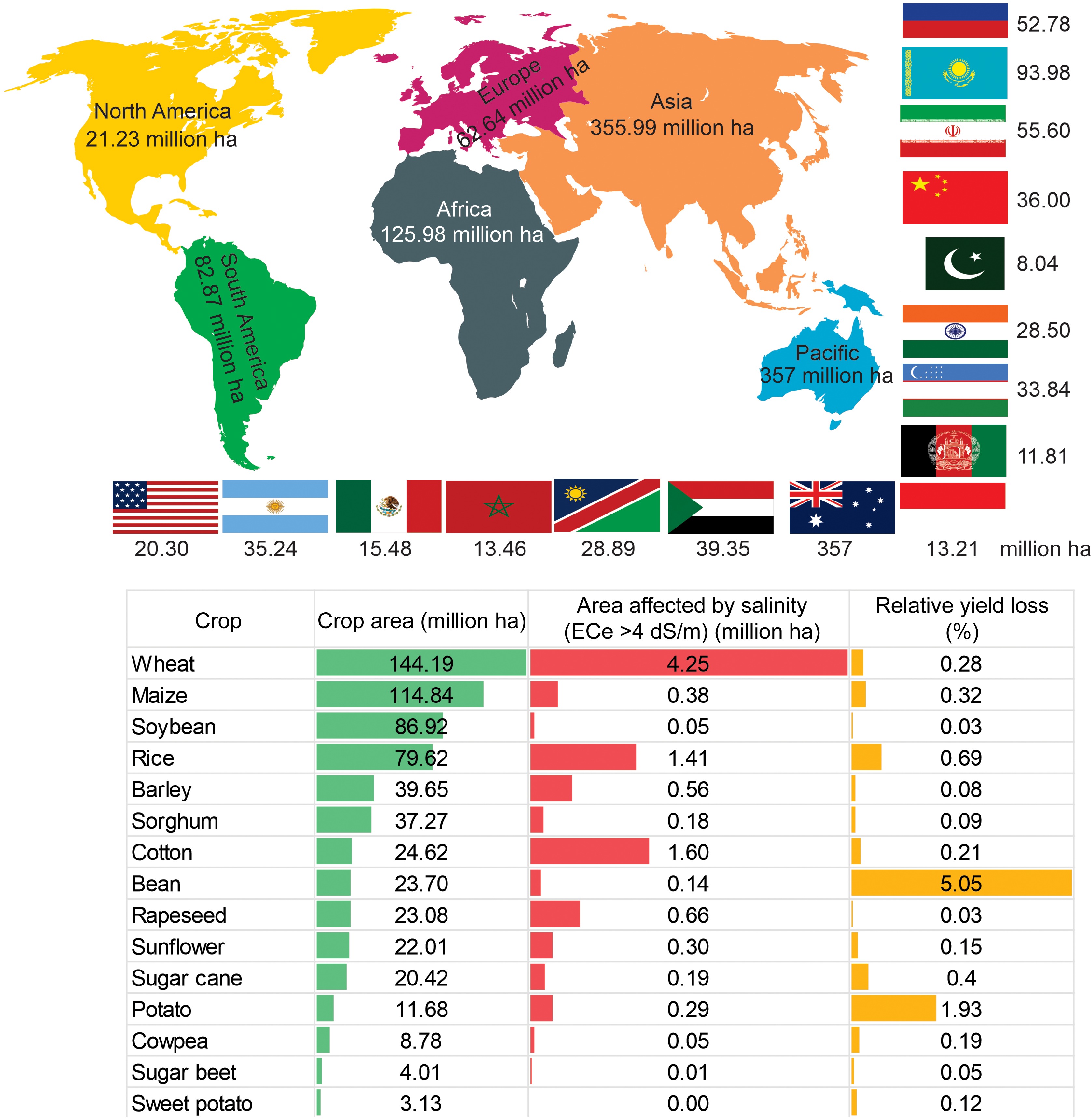
Figure 1.
Global map of the distribution of salt-affected soil in six continents and major countries affected by salt in each continent (million ha) (upper panel). Relative yield loss due to salinity in specific crops (lower panel). Global crop area (million ha) for each crop cultivation, area affected by salinity (million ha), and percent relative yield loss for each crop (%). The data on the global status of salt-affected soils were sourced from FAO 2024[11].
-
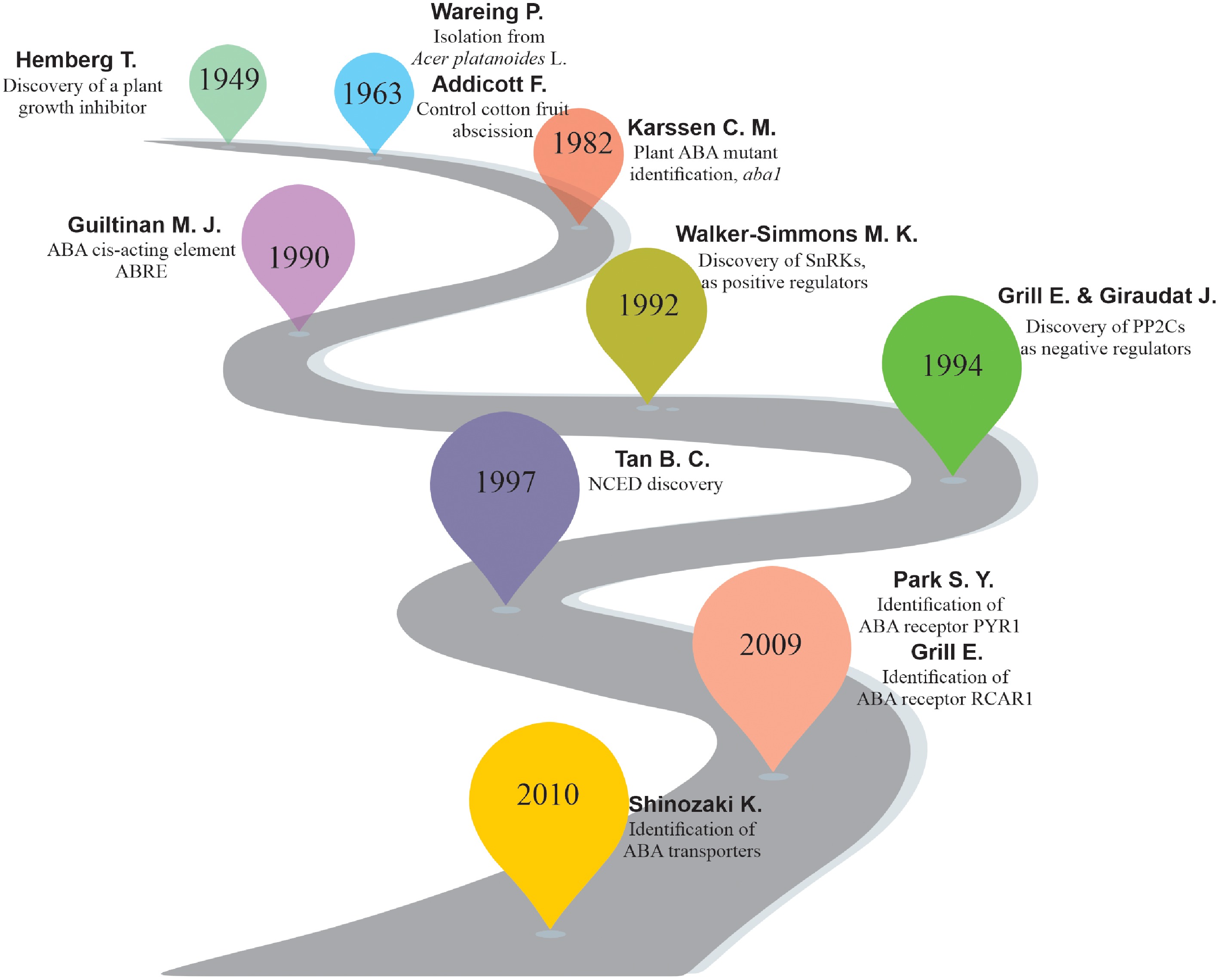
Figure 2.
Brief overview of ABA research progress. The complexity and diversity of the ABA pathway have been revealed through genetic, proteomic, and genomic approaches to elucidate the functions of ABA in agricultural production and plant biology research.
-
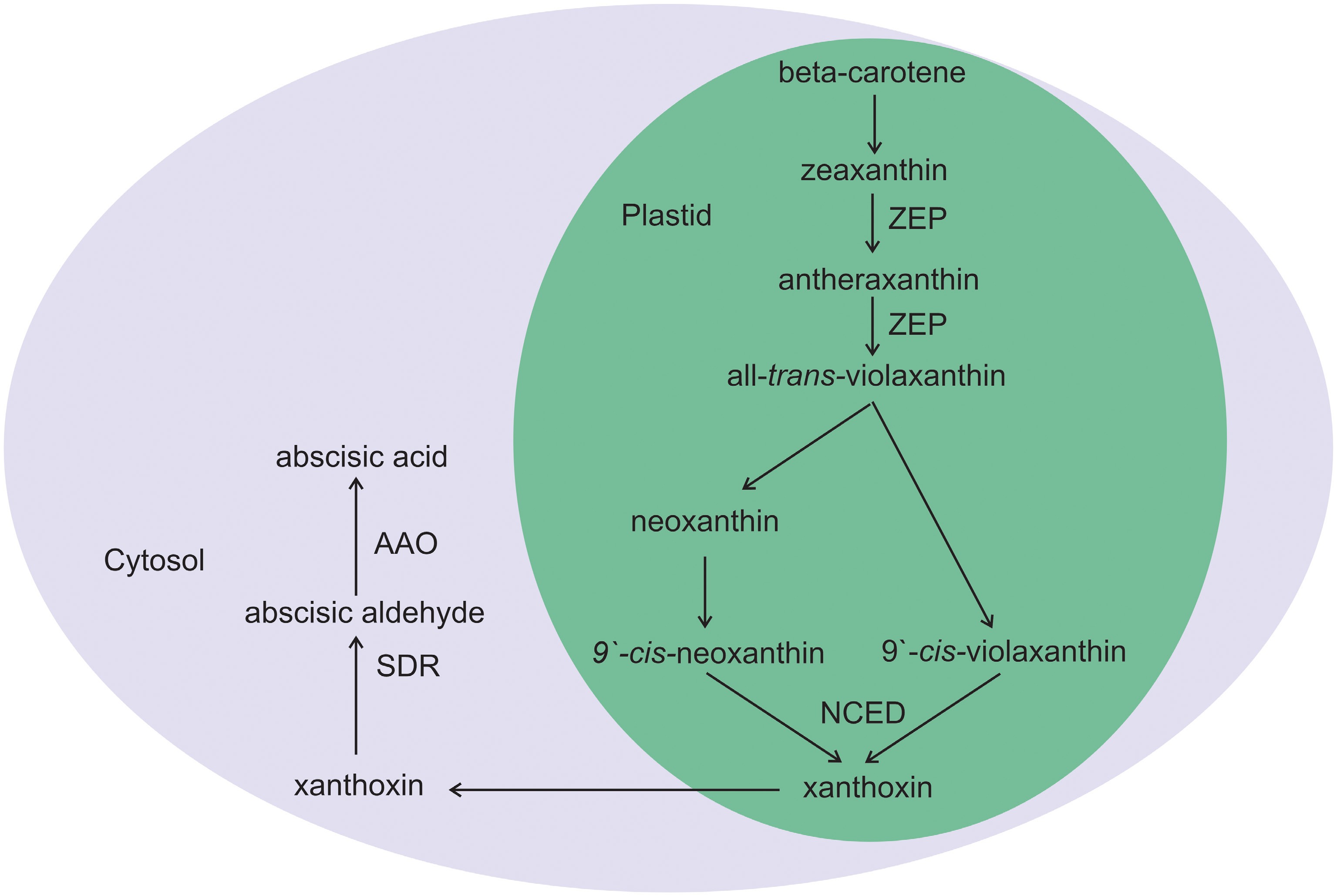
Figure 3.
Schematic diagram of ABA biosynthesis in plants. ABA biosynthesis occurs in plastids and involves different steps that are cleaved into xanthoxin derived from beta-carotenoid precursors. Xanthoxin is then exported to the cytosol from the plastid to form the final product ABA through two-step enzymatic catalysis by short-chain alcohol dehydrogenase/reductase (SDR) and abscisic aldehyde oxidase (AAO). Other enzymes involved in ABA biosynthesis are zeaxanthin epoxidase (ZEP) and 9-cis-epoxycarotenoid dioxygenase (NCED), which are located in the plastid.
-
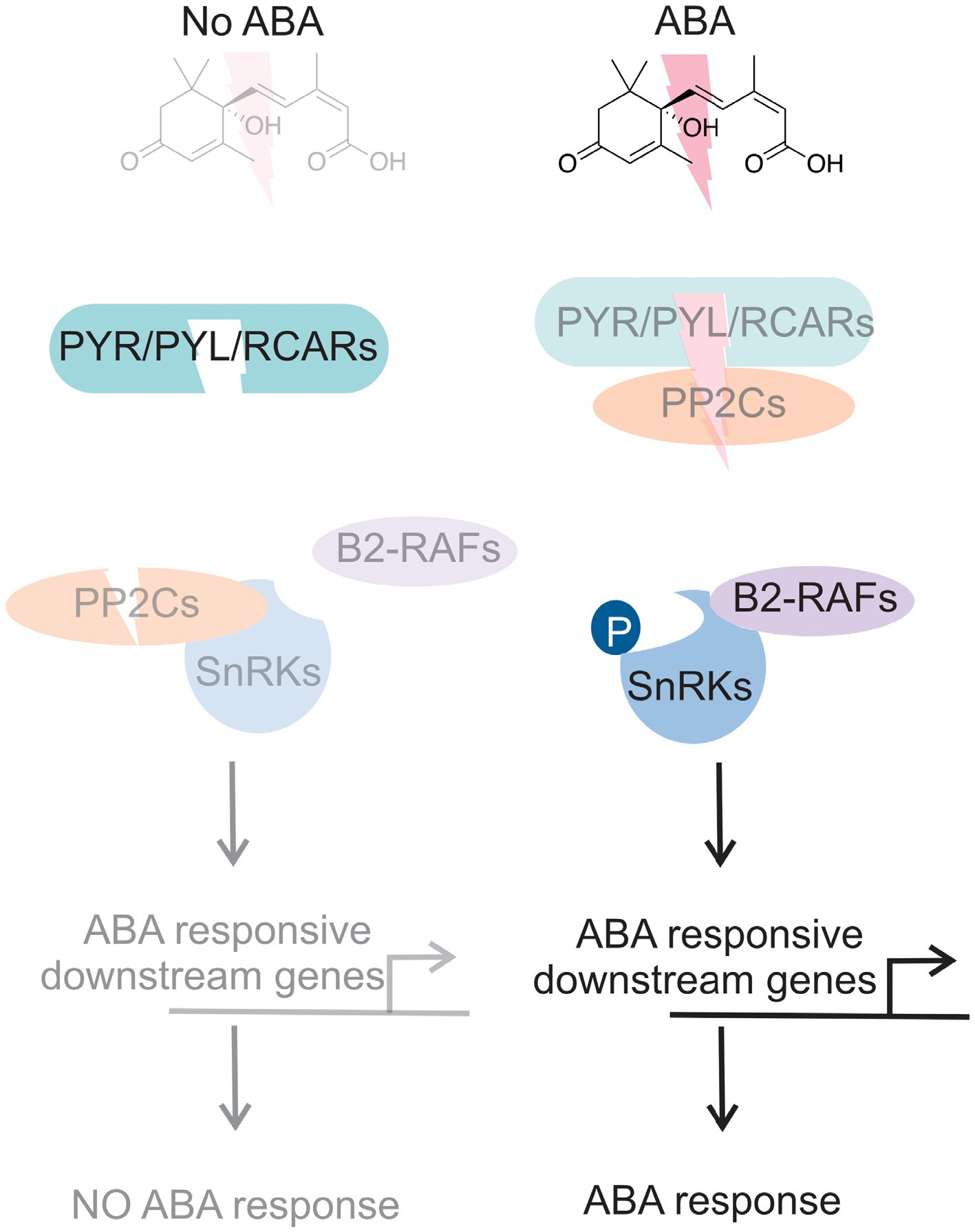
Figure 4.
ABA signaling model. As shown in the left panel, in the absence of ABA, Pyrabactin Resistance (PYR)/PYR1-like (PYL)/Regulatory Component of ABA Receptor (RCAR) family inactivated to activate ABA receptor proteins, a protein phosphatase 2C (PP2C), to inhibit the activity of subclass III sucrose nonfermenting 1-related protein kinases (SnRK) resulted in no ABA-mediated response. In the presence of ABA, PYR/PYL/RCAR bind to PP2C, releasing SnRKs from inhibition. Auto-activation allows the phosphorylation of downstream ABA-responsive genes, leading to ABA-mediated responses. Light color indicates inactivation, whereas dark color indicates activation.
-
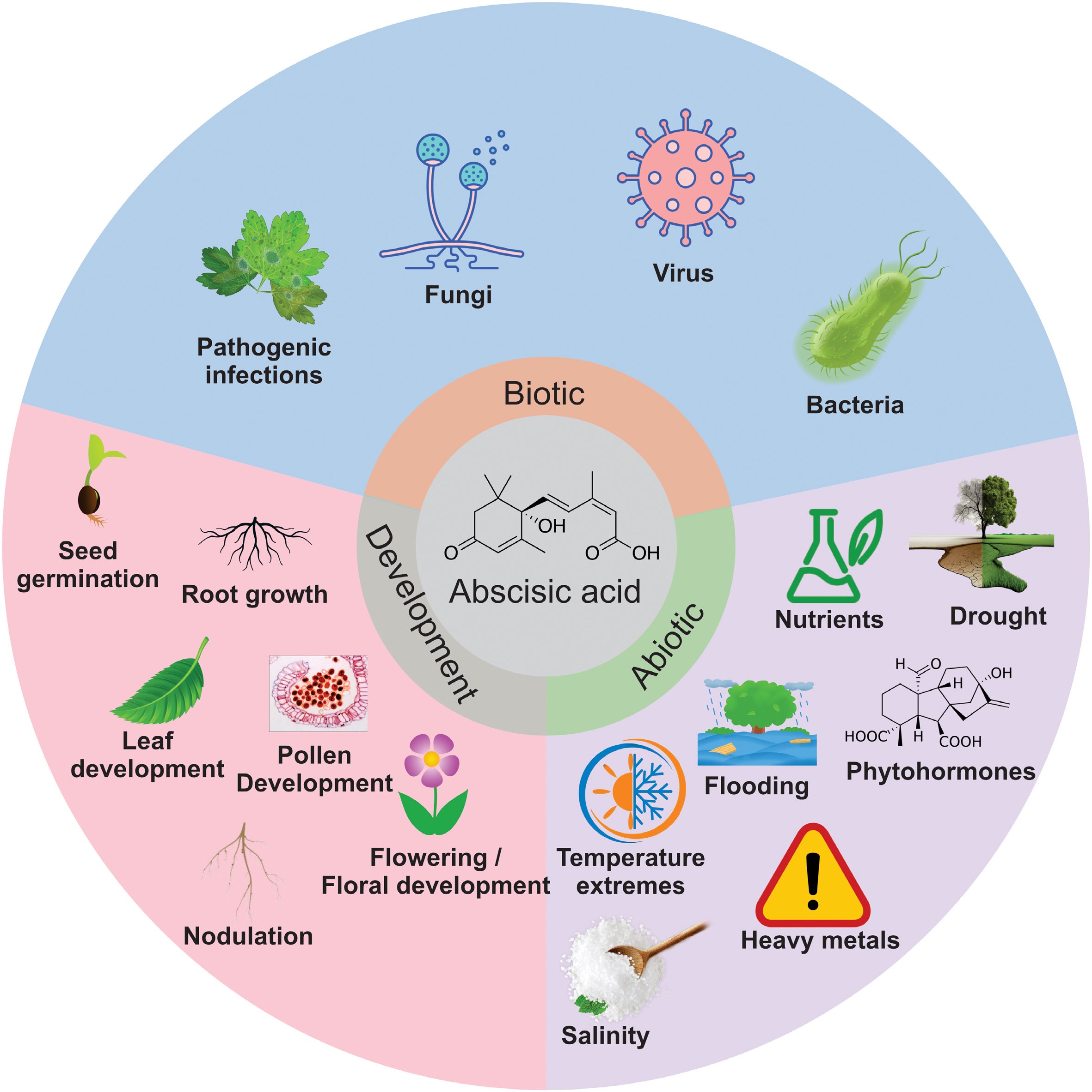
Figure 5.
Active regulation of a wide array of abiotic and biotic responses in plants by abscisic acid (ABA).
-
Plant Gene/protein/
transporterBiological function Ref. Rice ABIL2 and SAPK10 Enhance ABA-signaling for ABA accumulation under salinity stress [29] NAC016 ABA-mediated salinity tolerance [53] WRKY50 As transcriptional repressor of NCED5 and mediated ABA-dependent seedling germination [54] bHLH38 ABA-dependent salt tolerance [55] ERF922 Induce ABA-signaling under salinity stress [56] ASR6 ASR6 interact with NCED1 to enhance ABA accumulation to improve salt and oxidative injuries caused [57] ATAF1 Elevated ABA levels enhance salinity tolerance [58] bZIP42 ABA-responsive Rab16 and LEA genes to confer stress tolerance [59] NAC3 Salinity tolerance [60] ABF2 Positive regulator of ABA-signaling under salt stress [61] bZIP23 Confers ABA sensitivity and salinity tolerance [62] Arabidopsis USP17 Salinity tolerance [63] SDH Decreased ABA sensitivity [64] bZIP36/37/38 ABA dependent salinity tolerance [65] NADK2 Regulator of ABA-mediated stomatal closure, [66] Apple DREB1 ABA dependent and independent salt tolerance [67] Cotton PLATZ1 Reduced salinity tolerance [68] WRKY6-like ABA-mediated salt tolerance and activation of ROS enzymes [69] WRKY17 Activate ABA-signaling and alteration of ROS production [70] Tomato AITR3 ABA-mediated salinity tolerance [71] bHLH22 Salinity tolerance [72] Grapes bHLH1 Salinity tolerance by ABA induced ROS scavenging and osmotic balance [73] Wheat CHP ABA-mediated downregulation enhances salt tolerance [74] Pongamia AITR1 ABA-mediated Salt tolerance [75] Table 1.
Genes involved in ABA-mediated salinity tolerance in various plants.
Figures
(5)
Tables
(1)