-
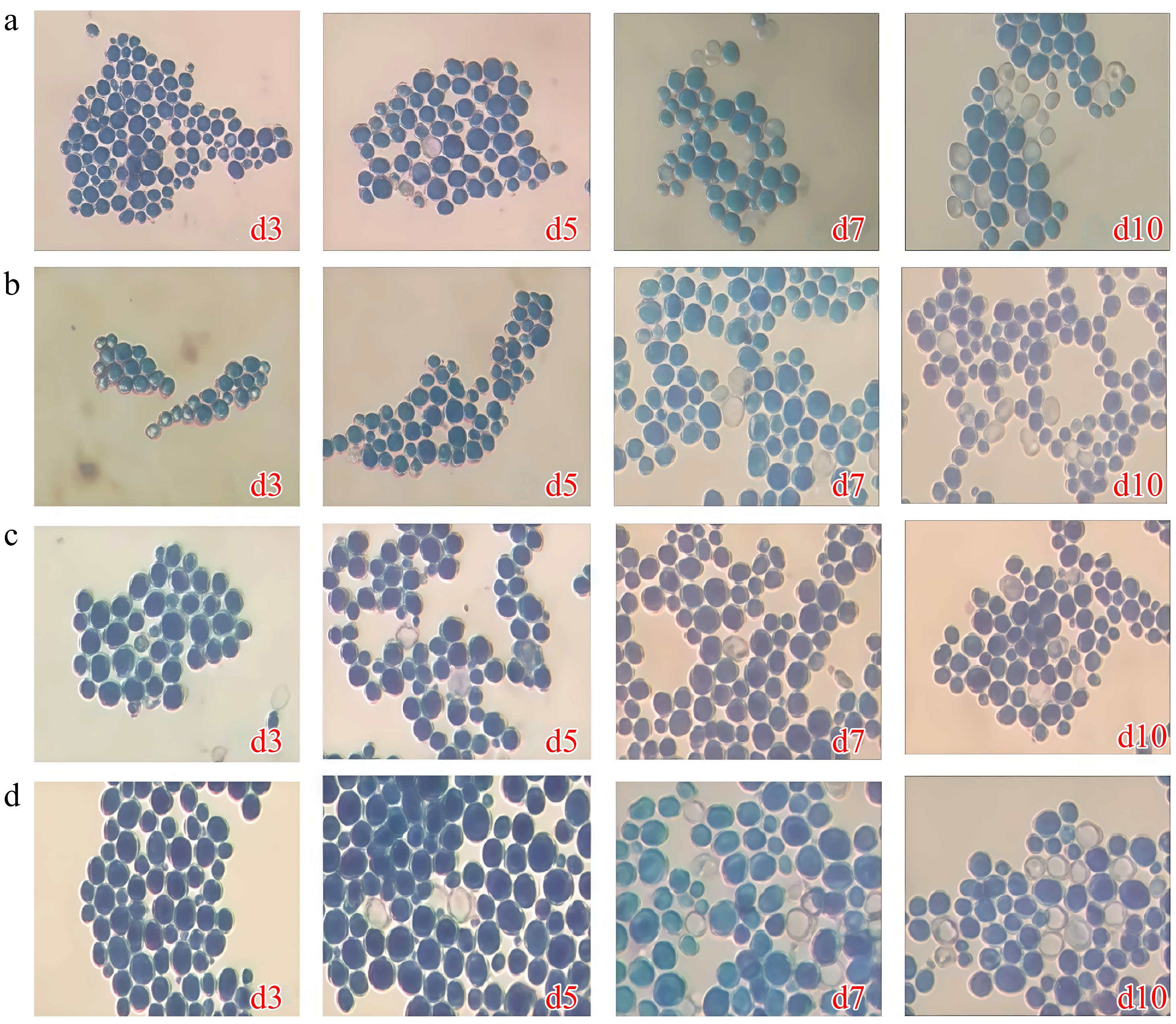
Figure 1.
Microscopic examination of S. cerevisiae 31y3 with methylene blue staining (× 400). (a) Kleyn's medium. (b) McClary medium. (c) SPM medium. (d) ACK medium.
-
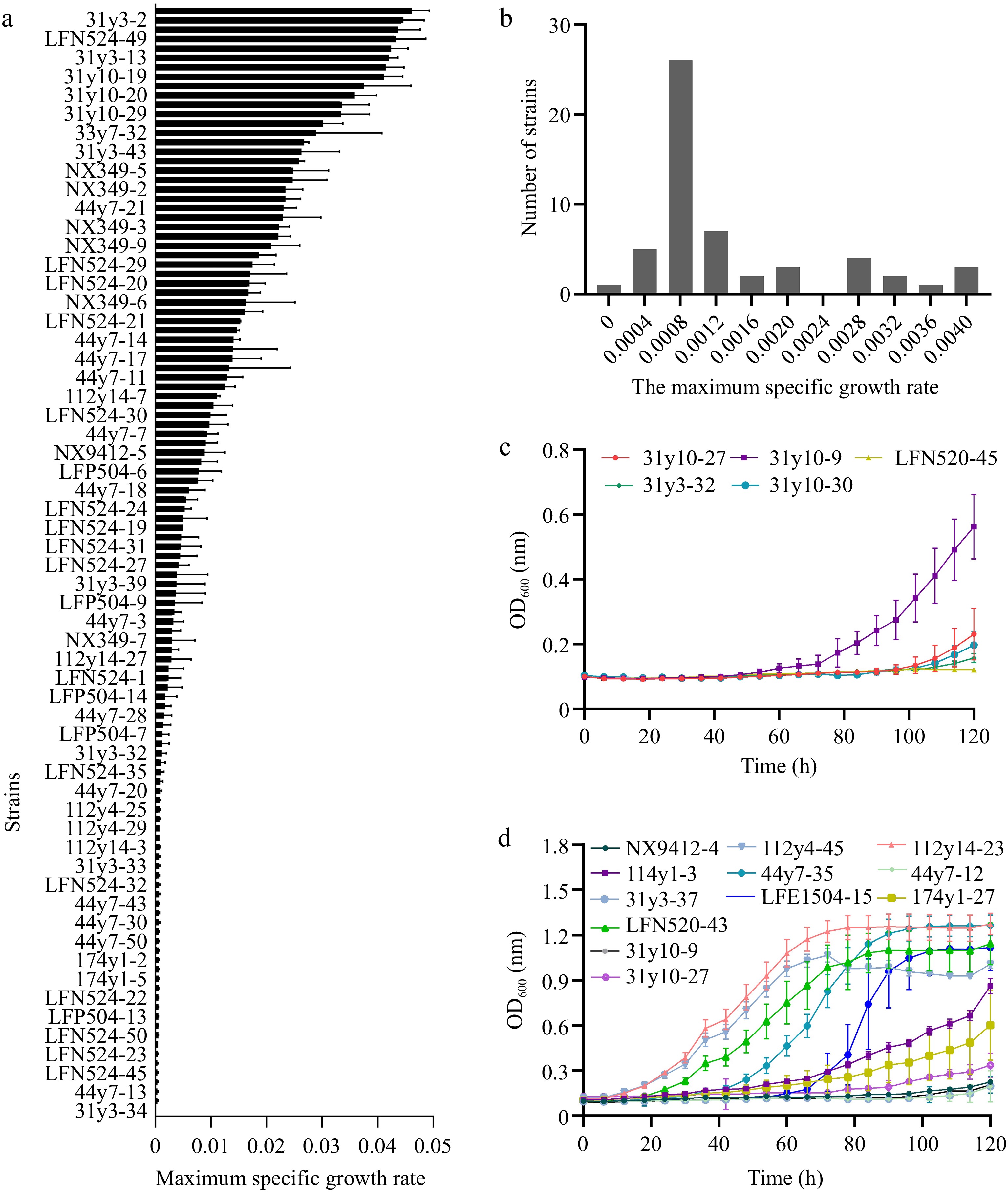
Figure 2.
Selection of haploid S. cerevisiae strains with different growth under combined ethanol-osmotic-SO2 stressors. (a) Maximum specific growth rate under combined ethanol-osmotic-SO2 stressors of the parental haploids. (b) Distribution of μmax in the stress-sensitive haploid S. cerevisiae strains. (c) Growth of stress-sensitive haploids in high-stress medium. (d) Growth of stress-tolerant haploids in low-stress medium.
-
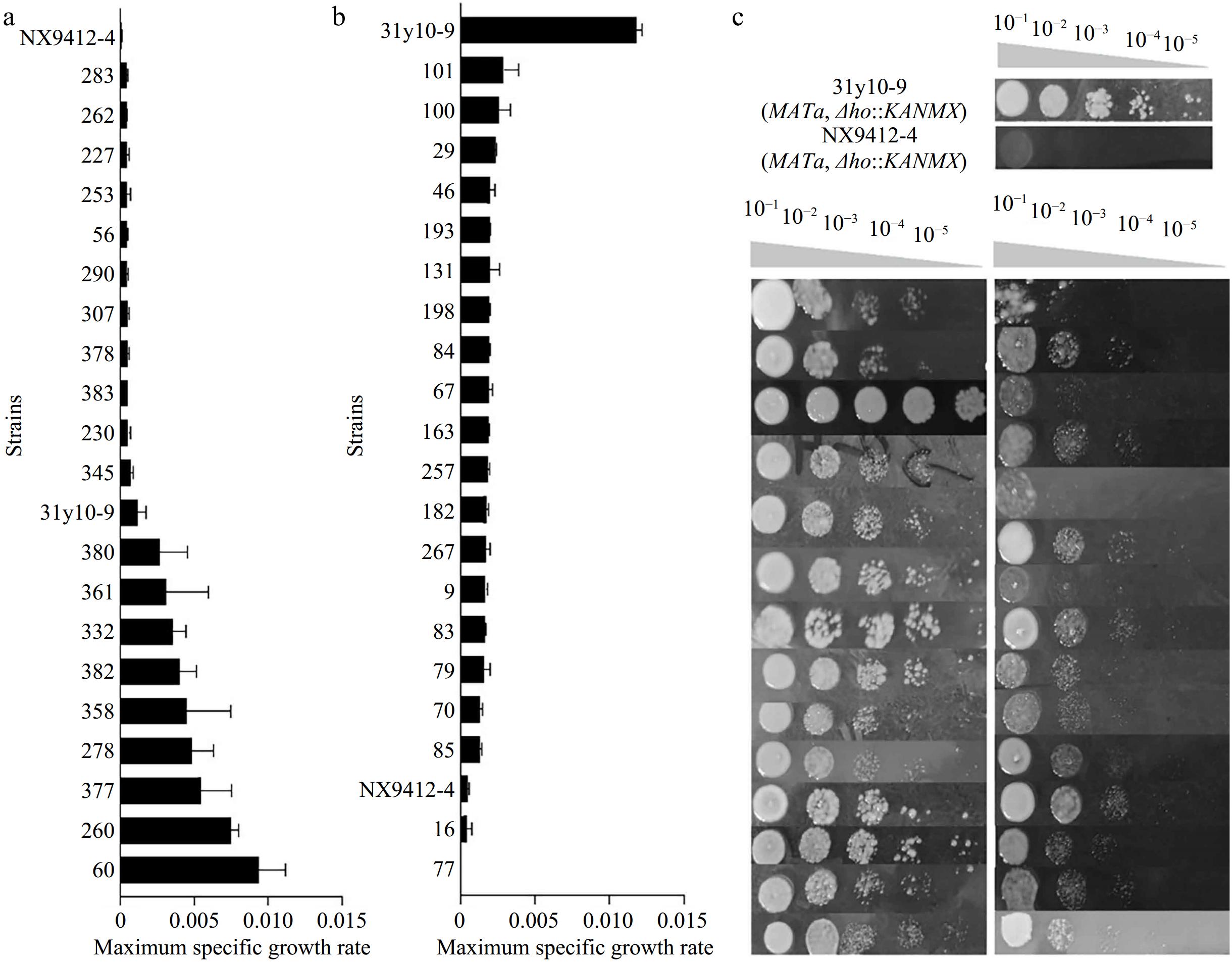
Figure 3.
The distribution of the μmax in the F2 segregants and construction of the two extreme pools. (a) Re-screening of the high-tolerance extreme pool. (b) Re-screening of the low tolerance extreme pool. (c) Spot growth of the superior segregants and inferior segregants from the two pools.
-
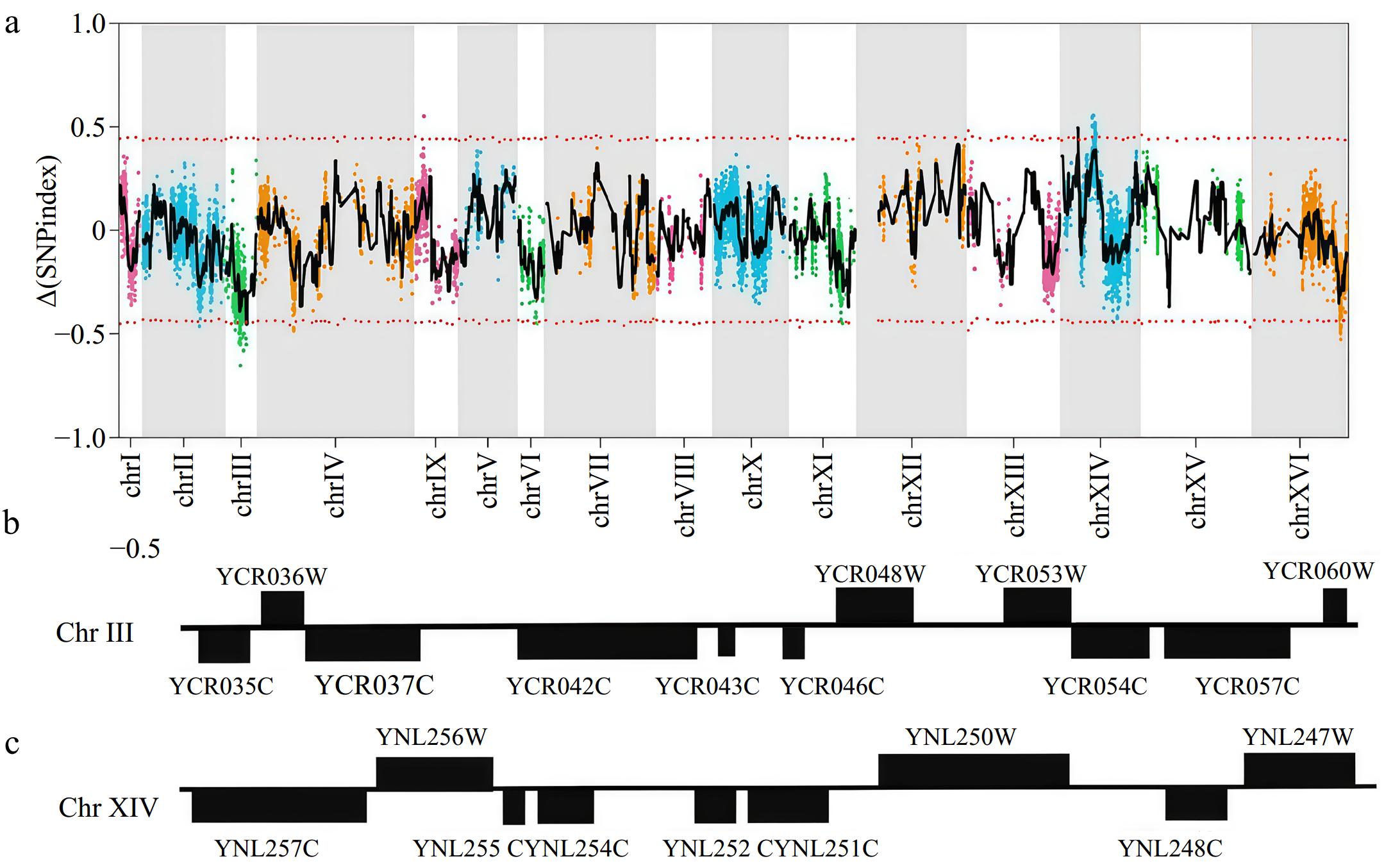
Figure 4.
Identification of QTLs by whole-genome re-sequencing analysis. (a) Mapping of the QTLs associated with multi-stress tolerance by pooled-segregant whole-genome sequencing analysis. The X-axis indicates the position of the chromosomes, whilst the Y-axis shows the Δ(SNP-index) values. (b) Overview of genes in the QTL on Chromosome III. (c) Overview of genes in the QTL on Chromosome XIV.
-
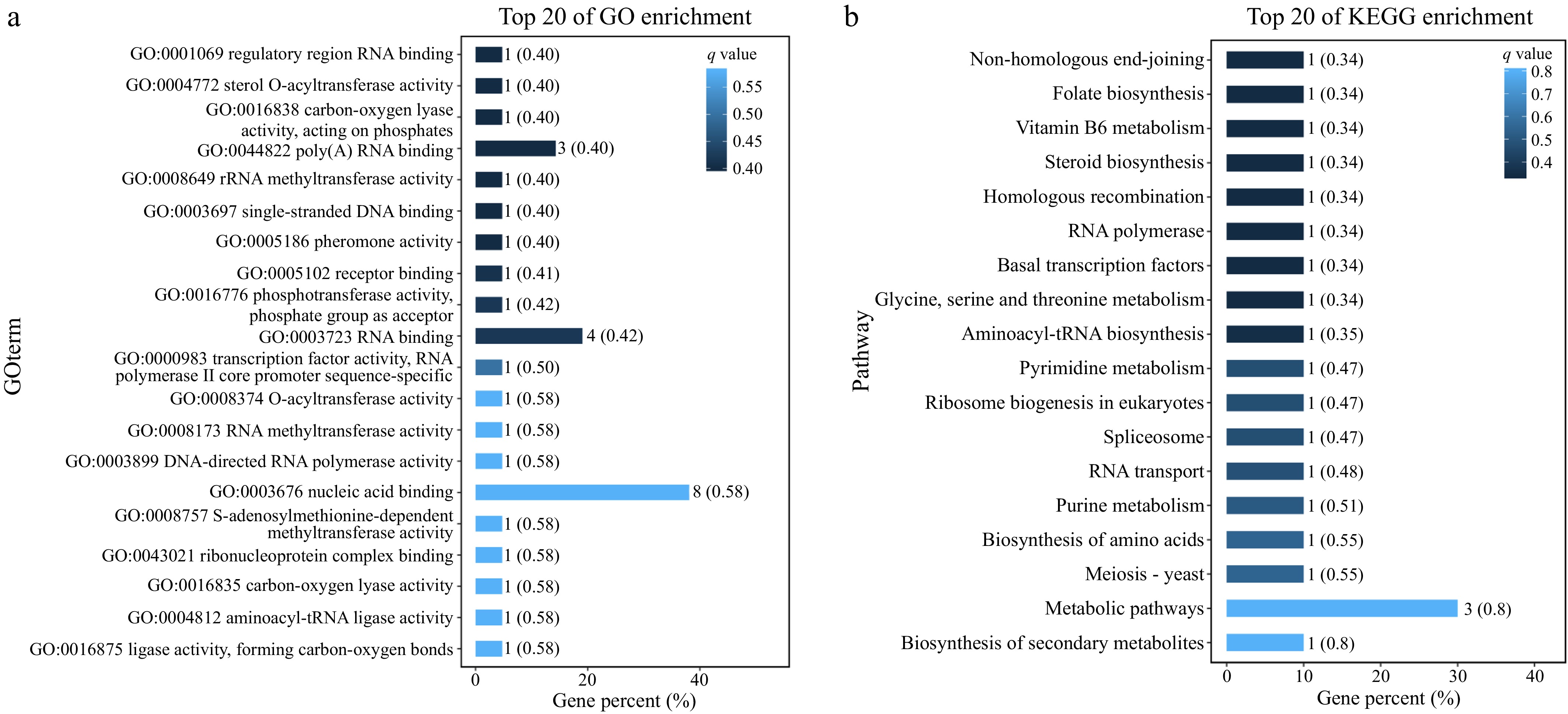
Figure 5.
(a) GO enrichment analysis of the genes in the two QTLs. (b) KEGG enrichment analysis of the genes in the two QTLs.
-
Classification of gene function Gene ID Name Gene functions RNA binding YNL251C NRD1 Regulatory region RNA binding; protein binding YCR057C PWP2 RNA binding YNL255C GIS2 Poly(A) RNA binding; transition metal ion binding DNA binding YNL250W RAD50 Phosphotransferase activity, phosphate group as acceptor; nucleoside-triphosphatase activity; purine ribonucleoside binding; cation binding; sequence-specific DNA binding rRNA methyltransferase activity YCR047C BUD23 rRNA methyltransferase activity sterol O-acyltransferase activity YCR048W ARE1 rRNA methyltransferase activity YCR053W THR4 Anion binding Table 1.
Candidate genes and their functions.
Figures
(5)
Tables
(1)