-
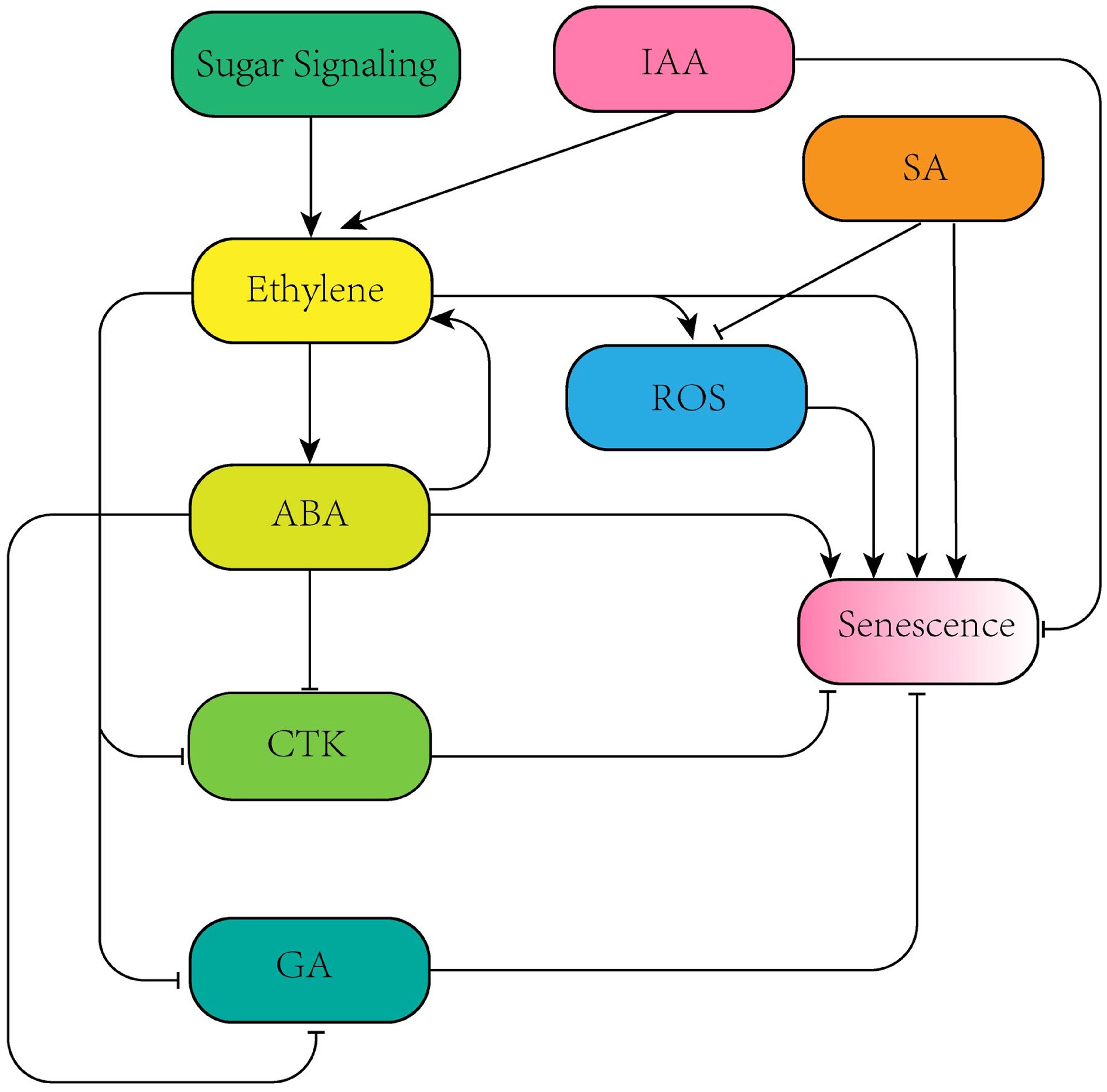
Figure 1.
Network diagram of synergistic and antagonistic effects among different hormones.
-
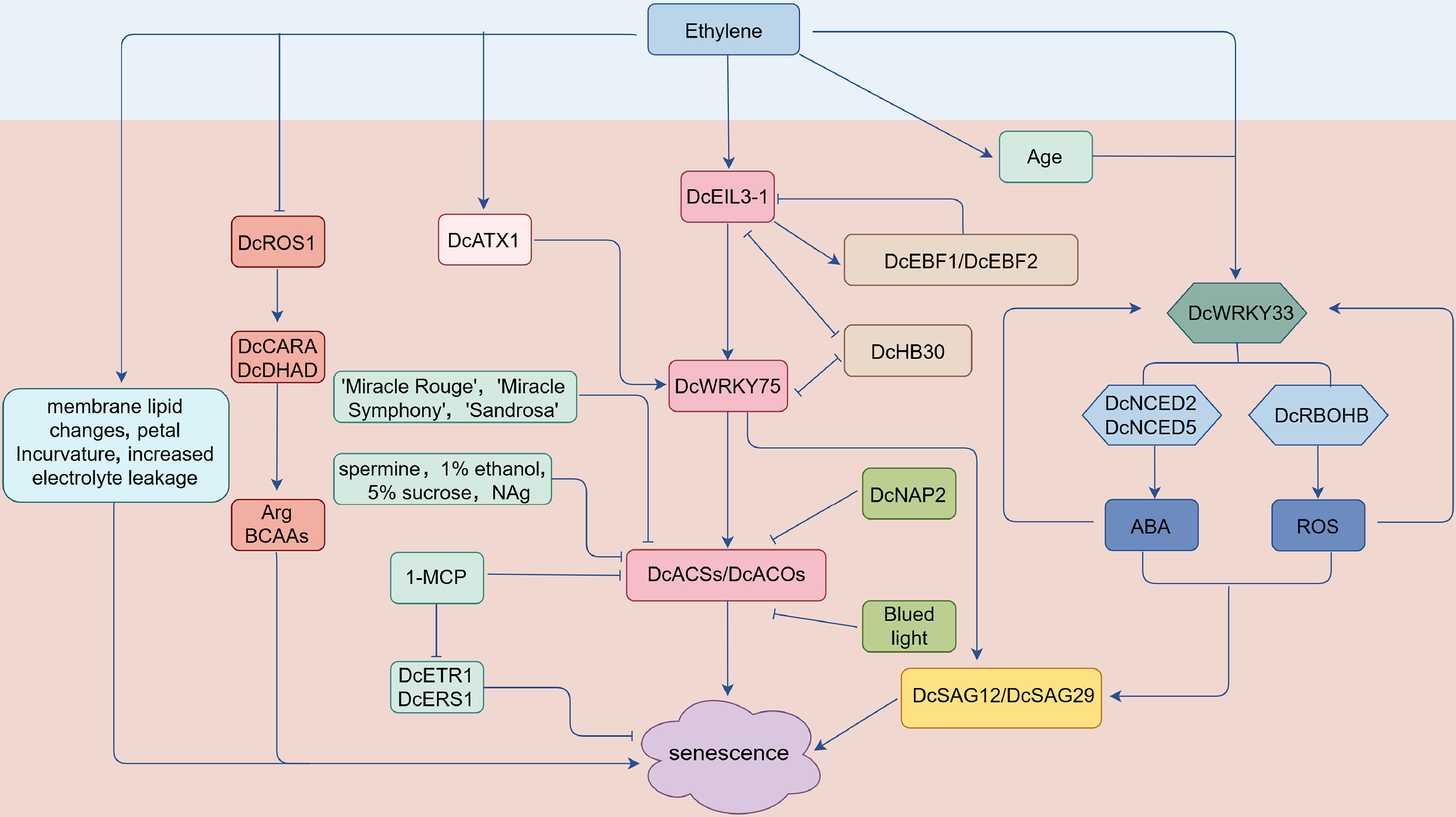
Figure 2.
Regulatory network diagram of ethylene-mediated petal senescence in carnation. ACS:1-aminocyclopropane-1-carboxylic acid (ACC) synthase; ACO: ACC oxidase; DcEIL3-1: core ethylene-responsive transcription factor (homolog of ethylene-insensitive 3-like 1); DcEBF1/2: homologs of EIN3-binding F-Box protein 1/2; DcROS1: DNA demethylase; DcATX1: histone methyltransferase; DcWRKY33, DcWRKY75: WRKY transcription factors; DcNCED2/5: 9-cis-epoxycarotenoid dioxygenase genes; DcRBOHB: respiratory burst oxidase homolog B; ABA: abscisic acid; DcCARA: carbamoyl-phosphate synthase subunit A; DcDHAD: dihydroxyacid dehydratase; Arg: arginine; BCAAs: branch chain amino acids; DcETR1, DcERS1: ethylene receptors; DcSAG12/29: senescence-associated genes.
-
Regulatory factor Ethylene ABA Target plants Ethylene-sensitive cut flowers (carnations and roses) Ethylene-insensitive cut flowers (chrysanthemums and lilies); sometimes collaborates with ethylene in sensitive flowers Biosynthetic pathway Catalyzed by ACC synthase (ACS) and ACC oxidase (ACO): SAM → ACC → ethylene, with ACS as the rate-limiting step Mediated by 9-cis-epoxycarotenoid dioxygenase (NCED): carotenoids → ABA, with NCED as the key rate-limiting enzyme Signaling pathway EIN2-EILs signaling cascade: Ethylene binds to receptors (DcETR1) → inhibits CTR1 → activates EIN2 → EILs transcription factors regulate senescence genes ABA accumulation induced by NCED activates WRKY, HD-Zip and other transcription factors, triggering oxidative stress Key genes/proteins ACS/ACO genes (DcACS1, DcACO1), signaling components EIN2,
DcEIL3-1, transcription factors DcWRKY33, RhERF3NCED genes (DcNCED2, DcNCED5), transcription factors TgWRKY75, PlMYB308, oxidase DcRBOHB Hormone interactions Antagonizes cytokinin (CTK): promotes CTK O-glucosylation and degradation; synergizes with ABA: activates ABA biosynthesis
genes via DcWRKY33Synergizes with ethylene: activates ethylene biosynthesis genes via RhERF3; antagonizes gibberellin (GA): inhibits GA synthesis and promotes membrane permeability Applications in preservation technology 1-MCP (irreversibly binds to ethylene receptors), AVG (inhibits ACS activity), sucrose (suppresses ethylene biosynthesis gene expression) ABA antagonist pyrabactin (inhibits ABA signaling), GA3 (antagonizes ABA effect), sucrose (regulates water and carbon metabolism) Table 1.
Comparison of regulatory mechanisms of ethylene and ABA in cut flower senescence.
Figures
(2)
Tables
(1)