-
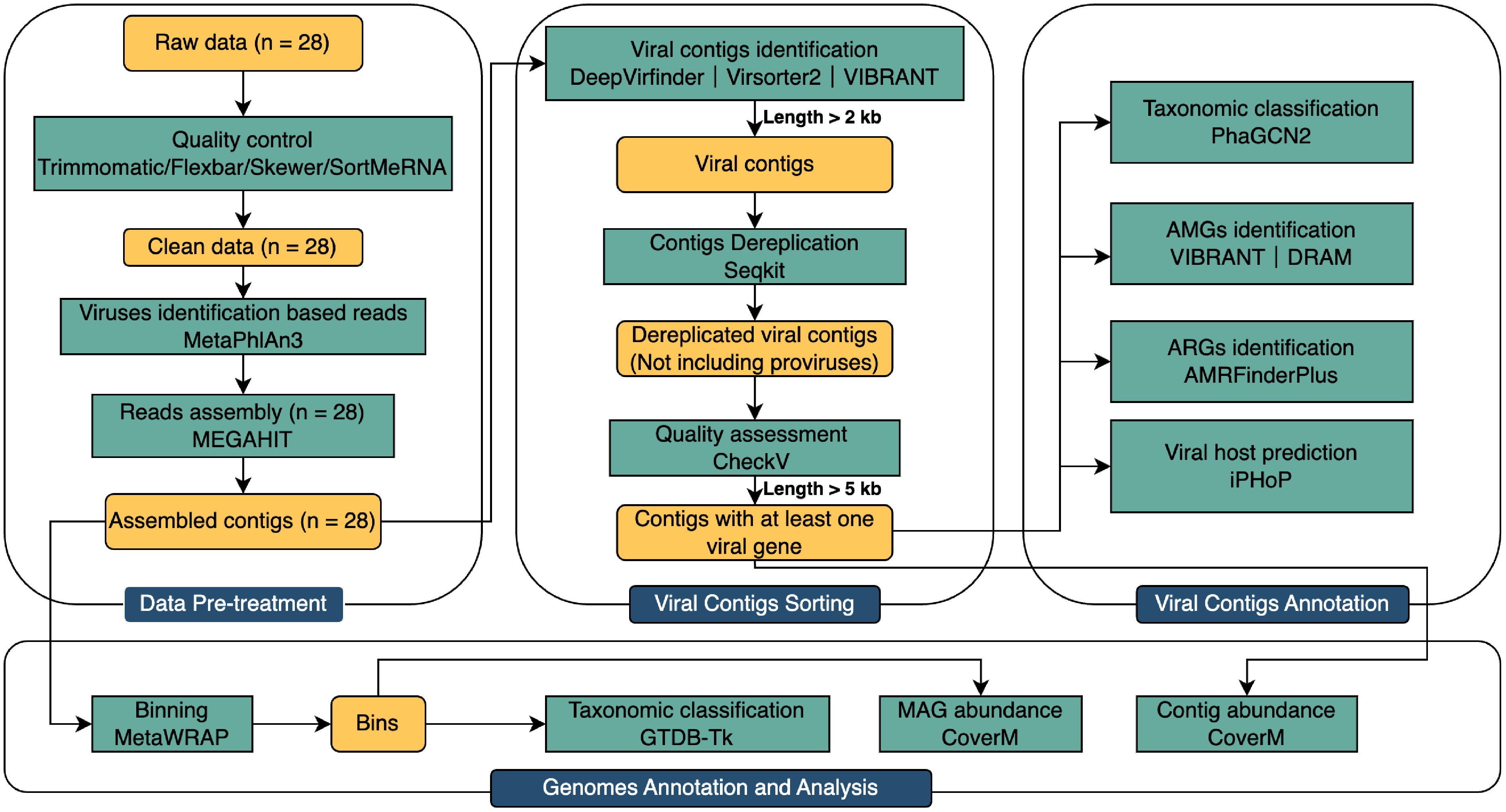
Figure 1.
Analytical diagram of the metagenomic virome data from raw data to binning.
-
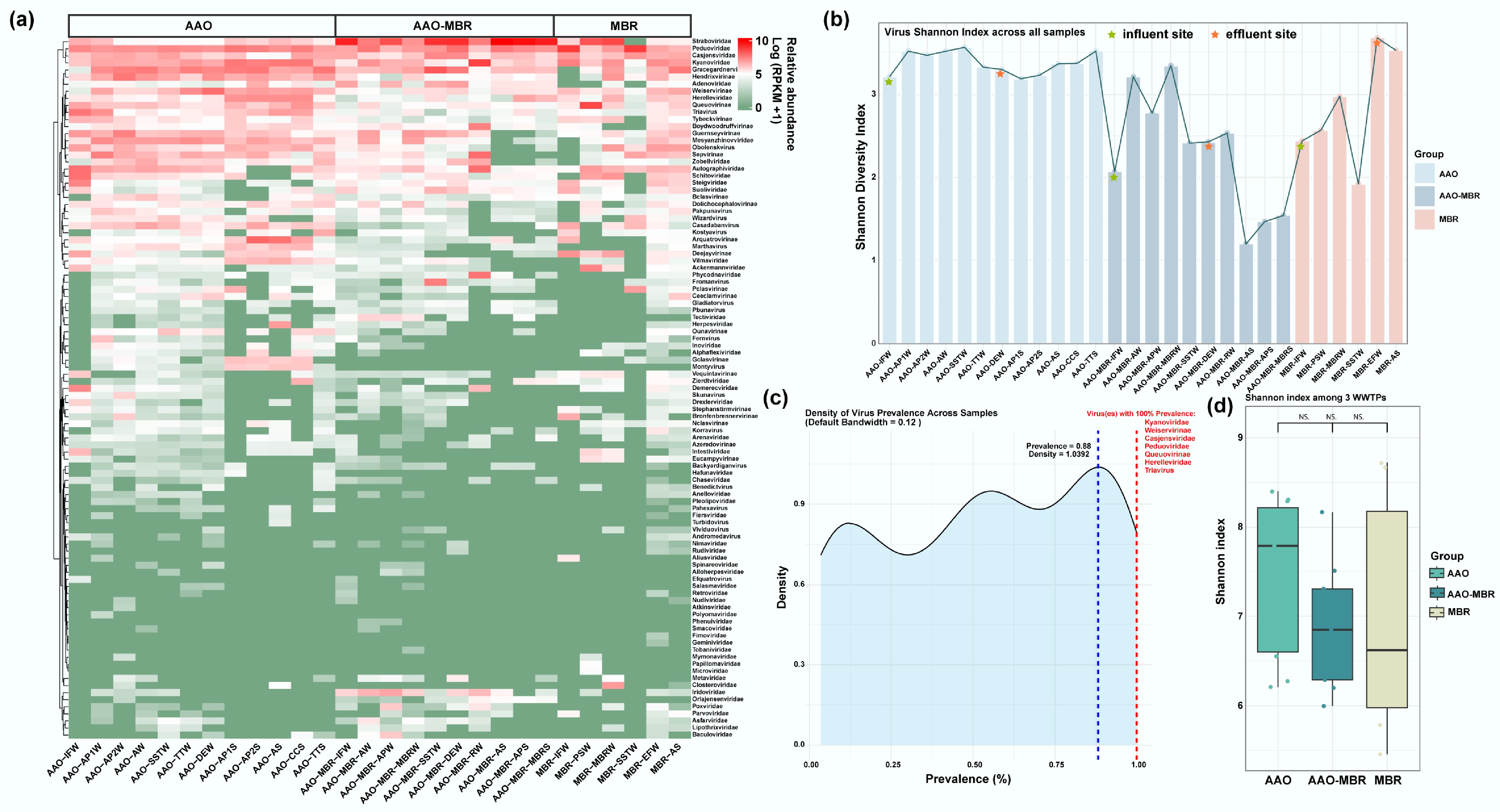
Figure 2.
Overview of virus distribution. (a) Relative abundance (log10 [RPKM + 1]) of identified viruses based on contigs along three full-scale WWTPs; (b) Shannon index of viruses across all samples. (c) Density curve of virus prevalence across samples (default bandwidth = 0.12), viruses denoted in red are persistent across full-scale with 100% prevalence. (d) Shannon diversity index among three WWTPs.
-
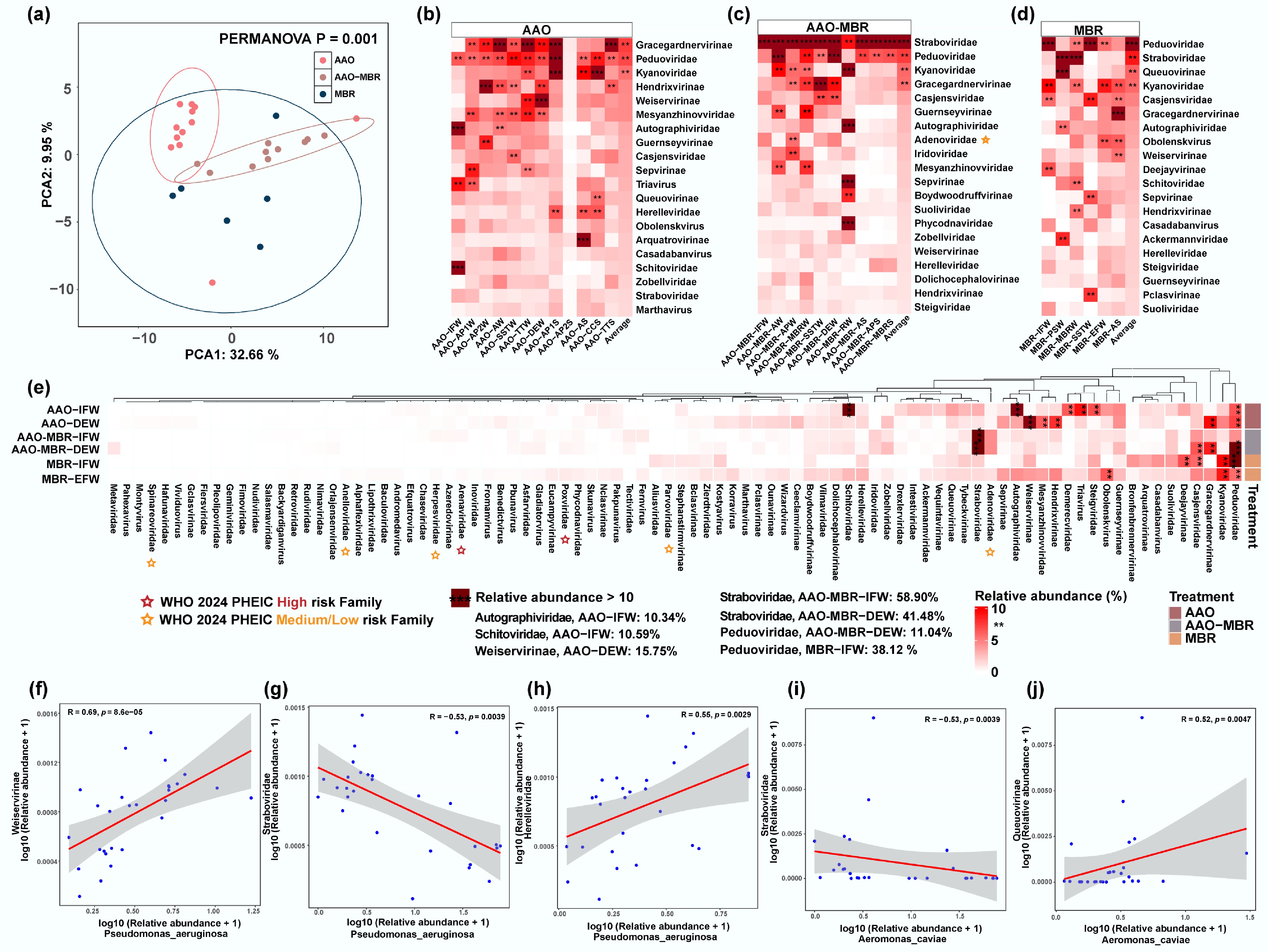
Figure 3.
(a) PCA analysis of viruses among three WWTPs. Composition and abundance of the top 20 most abundant (average abundance) viruses along the three full-scale WWTPs. (b) AAO WWTP. (c) AAO-MBR WWTP. (d) MBR. (e) A total of 88 viruses were detected in the influent and effluent of three WWTPs, and the abundance heatmap. Viruses with relative abundance > 10% were highlighted with ***, and ** refers to relative abundance between 5% and 10%. The risk family on the WHO 2024 PHEIC list was marked in red (high risk), and orange (medium/low risk). (f)–(j) Linear regression analyses between representative pathogen and virus with highest prevalence and abundance—(f)–(h) P. aeruginosa; (i), (j) A. caviae.
-
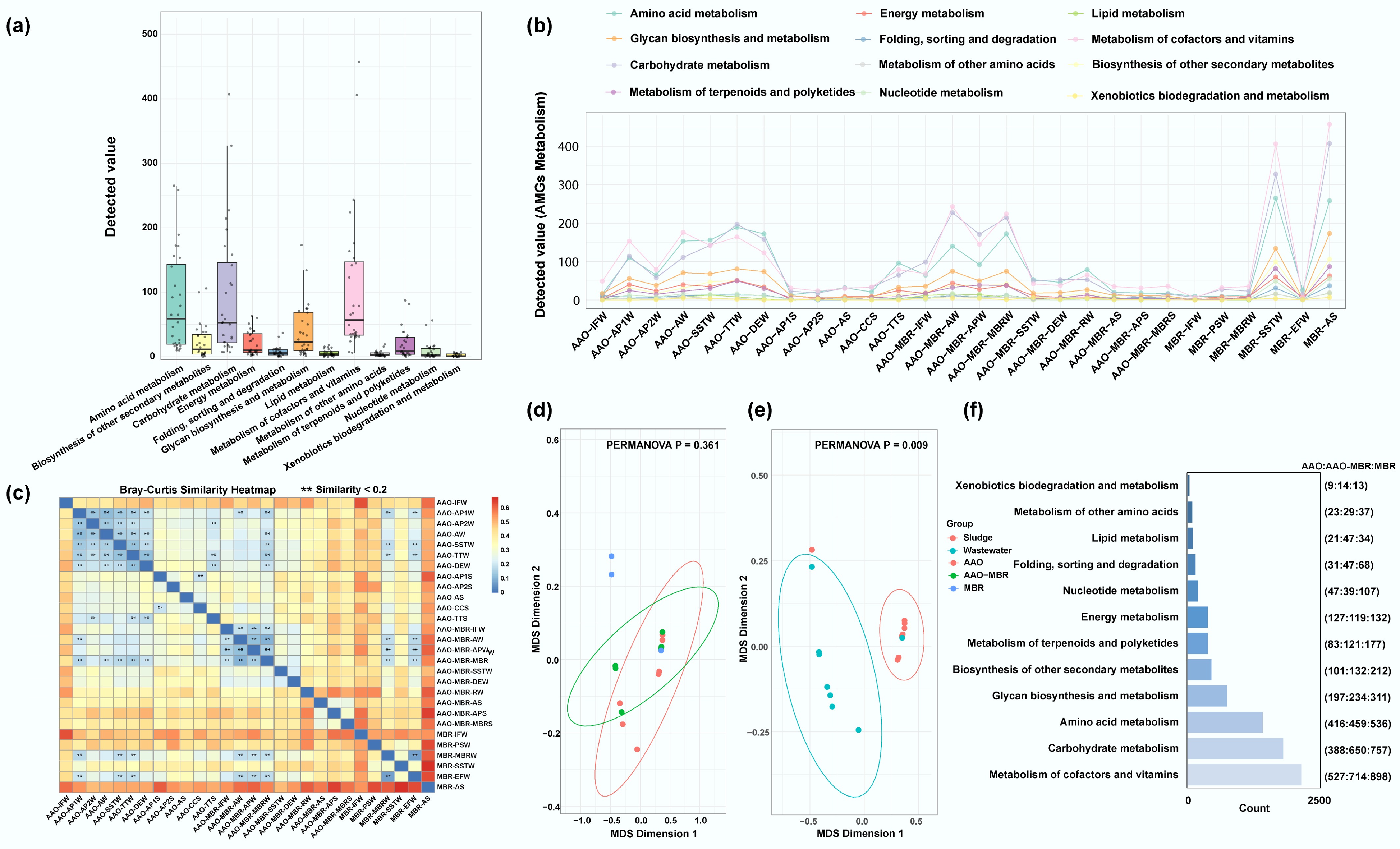
Figure 4.
Occurrence of AMGs and their full-scale regulation in three WWTPs. (a), (b) Detected 12 metabolism AMGs value across all samples and their distribution along the full-scale process, respectively. (c) Bray-Curtis similarity heatmap of AMGs across all the samples, ** refers to that similarity < 0.2. (d), (e) MDS analyses of AMGs in bioreactor sections among three WWTPs (ns. p > 0.05), and between wastewater and sludge (** p < 0.01), respectively. (f) The left bar plot represents the number of detected metabolism-related genes, while the numbers in parentheses indicate the number of corresponding AMGs identified in each WWTP.
-
Ranking AAO-IFW AAO-DEW AAO-MBR-IFW AAO-MBR-DEW MBR-IFW MBR-EFW 1 Schitoviridae *Weiservirinae *Straboviridae Straboviridae **Peduoviridae Kyanoviridae 2 Autographiviridae *Gracegardnervirinae Adenoviridae **Peduoviridae *Kyanoviridae Obolenskvirus 3 Triavirus *Hendrixvirinae **Peduoviridae *Gracegardnervirinae Deejayvirinae **Peduoviridae 4 *Steigviridae **Peduoviridae **Casjensviridae **Casjensviridae **Casjensviridae *Hendrixvirinae 5 **Peduoviridae Mesyanzhinovviridae Gracegardnervirinae Adenoviridae Casadabanvirus **Casjensviridae 6 Demerecviridae Kyanoviridae *Suoliviridae *Guernseyvirinae Bronfenbrennervirinae *Autographiviridae 7 Guernseyvirinae *Guernseyvirinae Iridoviridae Zobellviridae *Suoliviridae *Gracegardnervirinae 8 Obolenskvirus Sepvirinae Hendrixvirinae Suoliviridae Arquatrovirinae *Weiservirinae 9 Queuovirinae *Autographiviridae Autographiviridae *Weiservirinae *Steigviridae Sepvirinae 10 Deejayvirinae Ceeclamvirinae *Steigviridae Iridoviridae Weiservirinae *Mesyanzhinovviridae 11 *Straboviridae Zobellviridae Guernseyvirinae Steigviridae *Tybeckvirinae Steigviridae 12 *Tybeckvirinae **Casjensviridae *Kyanoviridae Schitoviridae Kostyavirus Herelleviridae 13 Intestiviridae Wizardvirus Zobellviridae Queuovirinae *Straboviridae Schitoviridae 14 Ackermannviridae Triavirus Obolenskvirus *Dolichocephalovirinae Herelleviridae *Guernseyvirinae 15 **Casjensviridae Ounavirinae Mesyanzhinovviridae Metaviridae Aliusviridae *Dolichocephalovirinae 16 *Kyanoviridae Herelleviridae *Tybeckvirinae *Mesyanzhinovviridae Queuovirinae Boydwoodruffvirinae 17 Vequintavirinae Queuovirinae Weiservirinae Vilmaviridae Stephanstirmvirinae Korravirus 18 *Suoliviridae *Dolichocephalovirinae Schitoviridae Boydwoodruffvirinae Parvoviridae Suoliviridae 19 Drexlerviridae Marthavirus Fernvirus *Hendrixvirinae Triavirus Tybeckvirinae 20 Arquatrovirinae Pclasvirinae Tectiviridae *Autographiviridae Boydwoodruffvirinae Vilmaviridae ** Peduoviridae, ** Casjensviridae are two of the top 20 viruses that persistently exist in six treatment units with the highest prevalence, marked in pink. Species marked in green and * are the viruses that persistently exist in influent sections, and species marked in grey and * are persistent in effluent. Table 1.
The top 20 relatively abundant viruses of influent and effluent in three WWTPs
-
AAO WWTP Contigs predicted AAO-MBR WWTP Contigs predicted MBR WWTP Contigs predicted *Acidobacteriota 18 *Acidobacteriota 14 *Acidobacteriota 9 *Actinomycetota 560 Actinomycetota 254 Actinomycetota 159 Aenigmatarchaeota 4 − − − − − − Armatimonadota 2 Armatimonadota 1 Asgardarchaeota 3 Asgardarchaeota 1 Asgardarchaeota 7 Atribacterota 4 Atribacterota 0 Atribacterota 0 *Bacillota 137 Bacillota 54 Bacillota 118 *Bacteroidota 356 *Bacteroidota 503 *Bacteroidota 297 − − − − Bipolaricaulota 1 Caldisericota 1 − − Caldisericota 3 *Campylobacterota 10 *Campylobacterota 2 *Campylobacterota 16 *Chloroflexota 88 *Chloroflexota 20 *Chloroflexota 24 *Cyanobacteriota 34 Cyanobacteriota 41 Cyanobacteriota 27 − − − − Deinococcota 2 *Dependentiae 3 *Dependentiae 6 Dependentiae 1 − − Desulfobacterota 2 *Desulfobacterota 5 Elusimicrobiota 5 − − − − Fibrobacterota 1 − − − − *Gemmatimonadota 20 Gemmatimonadota 1 Gemmatimonadota 11 − − Goldbacteria 0 Goldbacteria 1 *Halobacteriota 1 Halobacteriota 1 Halobacteriota 4 Hydrogenedentota 4 − − − − Iainarchaeota 1 − − − − Krumholzibacteriota 32 − − Krumholzibacteriota 3 Methanobacteriota 2 Methanobacteriota 16 Methanobacteriota 8 − − Methylomirabilota 1 − − − − − − Muirbacteria 2 *Myxococcota 93 *Myxococcota 56 Myxococcota 26 Nanoarchaeota 6 − − Nanoarchaeota 2 *Nitrospirota 26 Nitrospirota 22 Nitrospirota 12 Omnitrophota 15 − − Omnitrophota 4 *Patescibacteria 47 *Patescibacteria 35 *Patescibacteria 33 *Planctomycetota 67 Planctomycetota 23 Planctomycetota 26 *Pseudomonadota 1,675 Pseudomonadota 855 Pseudomonadota 637 Spirochaetota 4 − − *Spirochaetota 2 Sumerlaeota 1 − − Sumerlaeota 2 − − Thermoplasmatota 1 Thermoplasmatota 10 − − Thermoproteota 1 Thermoproteota 4 *Verrucomicrobiota 28 *Verrucomicrobiota 29 Verrucomicrobiota 17 Taxonomies identified from recovered MAGs are highlighted in red. Table 2.
The quantity of predicted host contigs across all sampling sites of three WWTPs
Figures
(4)
Tables
(2)