-

Figure 1.
Environmental surveillance and mitigation strategies for antimicrobial resistance.
-

Figure 2.
Phenotyping and genotyping approaches for environmental AMR surveillance.
-
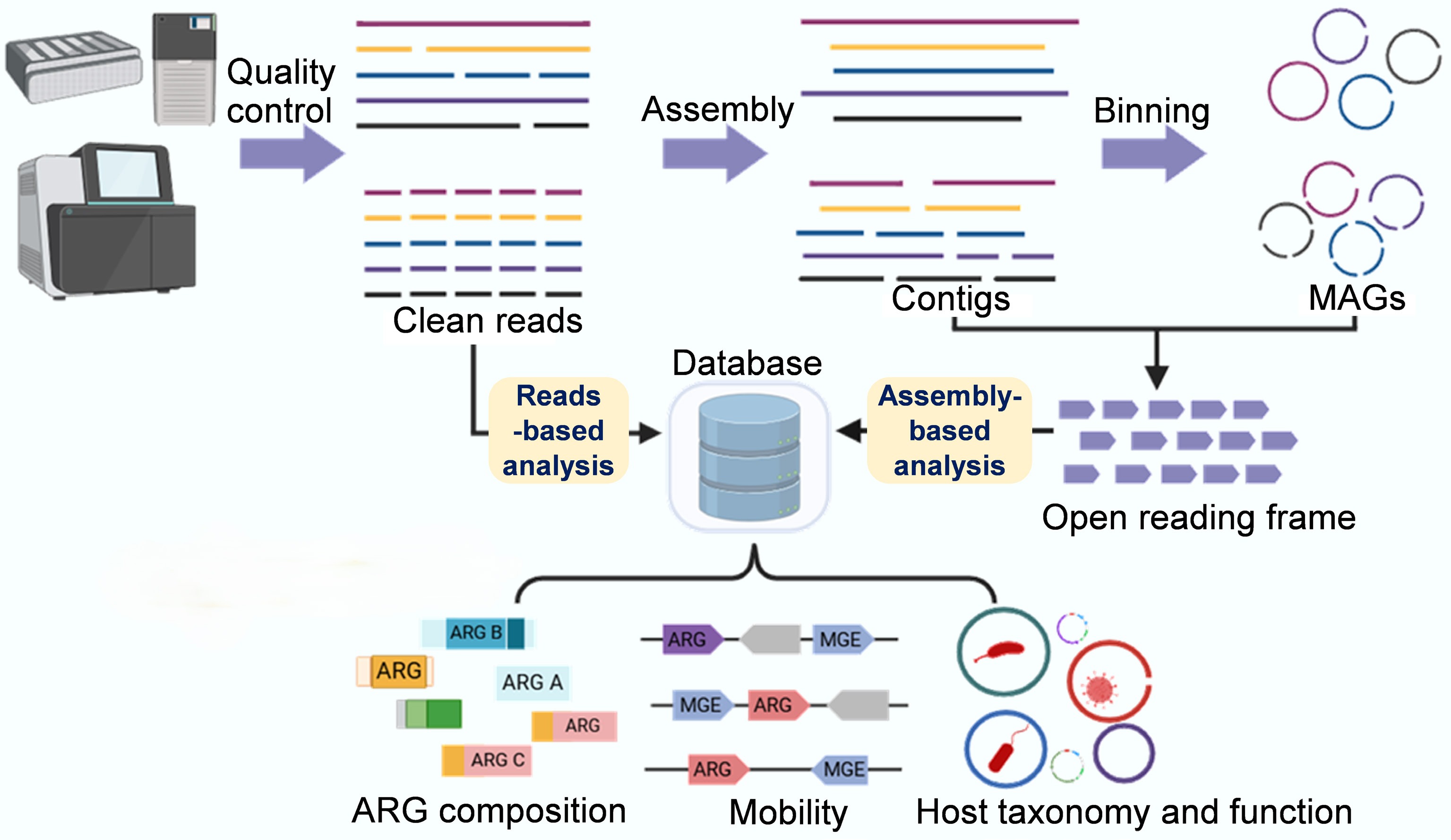
Figure 3.
Metagenomic sequencing-based analysis workflow of ARGs and resistomes. The basic traits of the resistome, including the diversity, abundance, and composition of ARGs, can be comprehensively analyzed using a short-read-based approach (which sensitively detects rare or hard-to-assemble species and genes). Key traits of the resistome, such as mobility, host pathogenicity, and multi-resistance, can be qualitatively analyzed using an assembly-based approach that produces metagenome-assembled genomes and longer contigs, thereby achieving refined taxonomic and phylogenetic resolution and improved functional insights.
-
Principles Advantages Limits Target Quantitative/ qualitative Ref. Phenotypic methods Culture-based Bacterial growth Standardized process; quantification Time-consuming; low throughout Isolator Quantitative [15,43,44] Flow cytometry Fluorescence counting Rapid; portable; single cell Small scale and volume Cell Quantitative [16] Raman spectroscopy Inelastic scattering or Raman scattering Rapid; label-free; single cell Limited bacterial species; low resolution Cell Qualitative [49−51] MALDI-TOF/MS Mass-to-charge ratio Rapid; label-free; high throughput High instrumental cost; culture-dependent Protein Qualitative [62−64] Genotypic methods PCR ARGs' conserved primer region High sensitivity; culture-free Unavailable information on resistome (ARG host, pathogenicity); restricted detection of unknown ARGs DNA Quantitative [68,69,72] CRISPR-Cas Cas protein cleavage High sensitivity/accuracy; Culture-free Limit bacterial species DNA/RNA Quantitative [17,83] Metagenomics ARG sequences High-throughput; information on the resistome (ARG host, pathogenicity); detection of new ARGs Requirement for expertise in bioinformatics; lack of standard protocol DNA Relative abundance [87,90] Integrated methods Phenotypic screening and genomics Comprehensive detection; enhanced accuracy; targeted surveillance Complex workflow; higher cost; technical expertise required Cell and DNA Quantitative/
qualitative[41,109,
111−113]Table 1.
Classic and emerging methods for quantitative and qualitative surveillance of antibiotic-resistant bacteria and genes
Figures
(3)
Tables
(1)