-
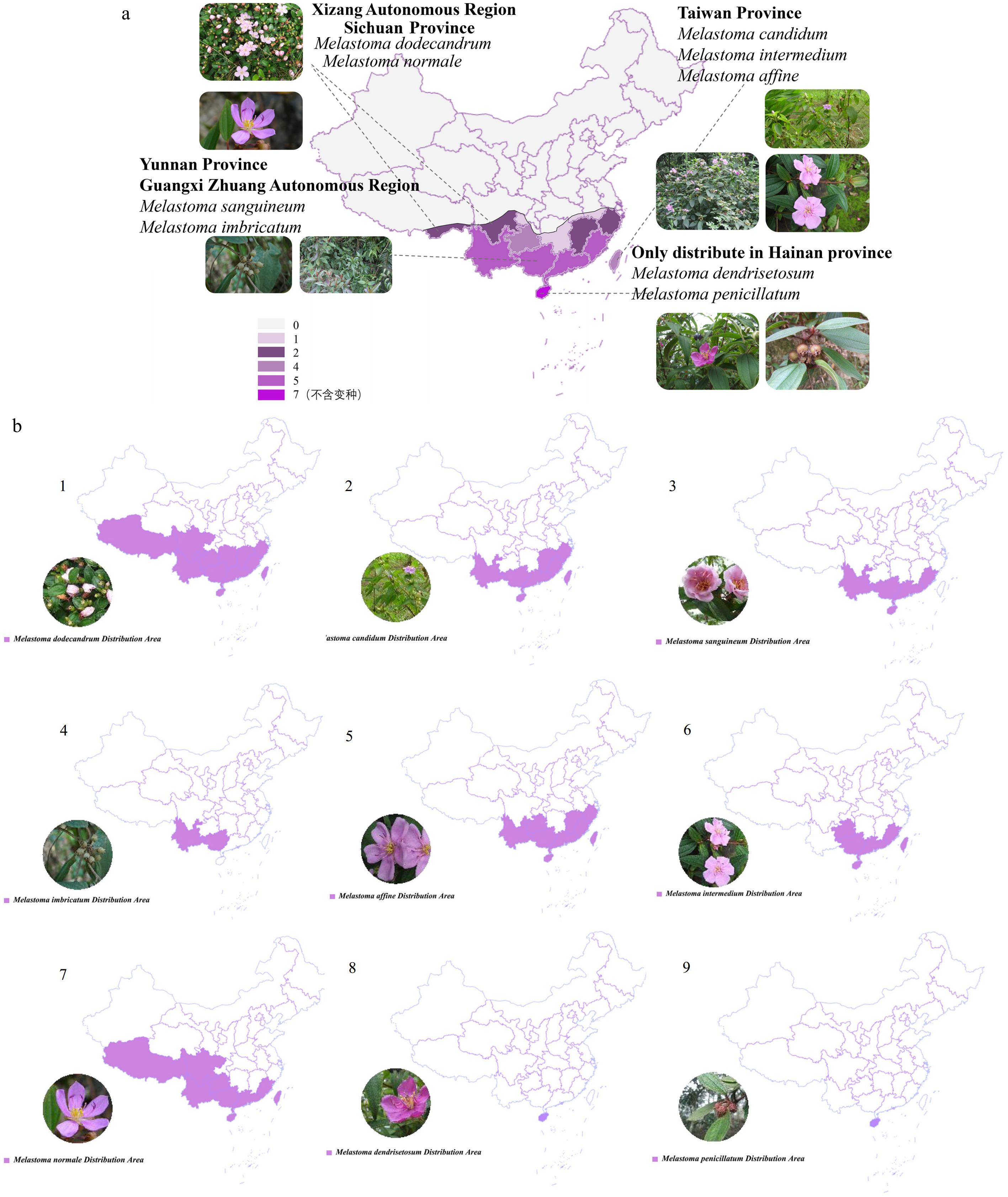
Figure 1.
Quantity distribution maps. (a) Species of Melastoma in various provinces of China. (b) The specific distribution of nine species of Melastoma plants. 1: Melastoma dodecandrum, 2: Melastoma candidum, 3: Melastoma sanguineum, 4: Melastoma imbricatum, 5: Melastoma affine, 6: Melastoma intermedium, 7: Melastoma normale, 8: Melastoma dendrisetosum, 9: Melastoma penicillatum.
-
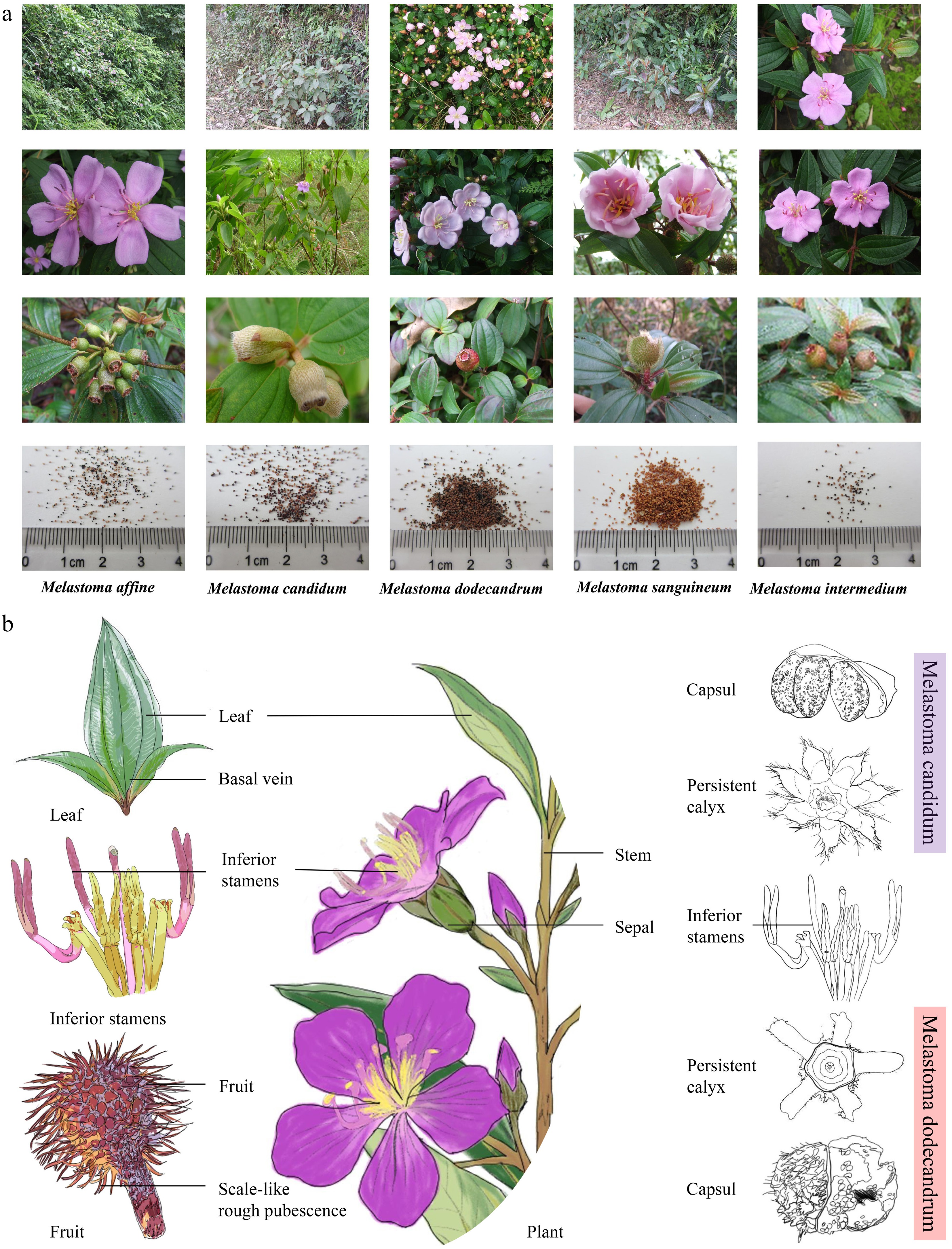
Figure 2.
(a) Characteristic images of five Melastoma species; the seed images were published in the monograph[19]. (b) Schematic diagram of plant structure of Melastoma represented by Melastoma dodecandrum and Melastoma candidum, including leaves, flowers, and fruits.
-
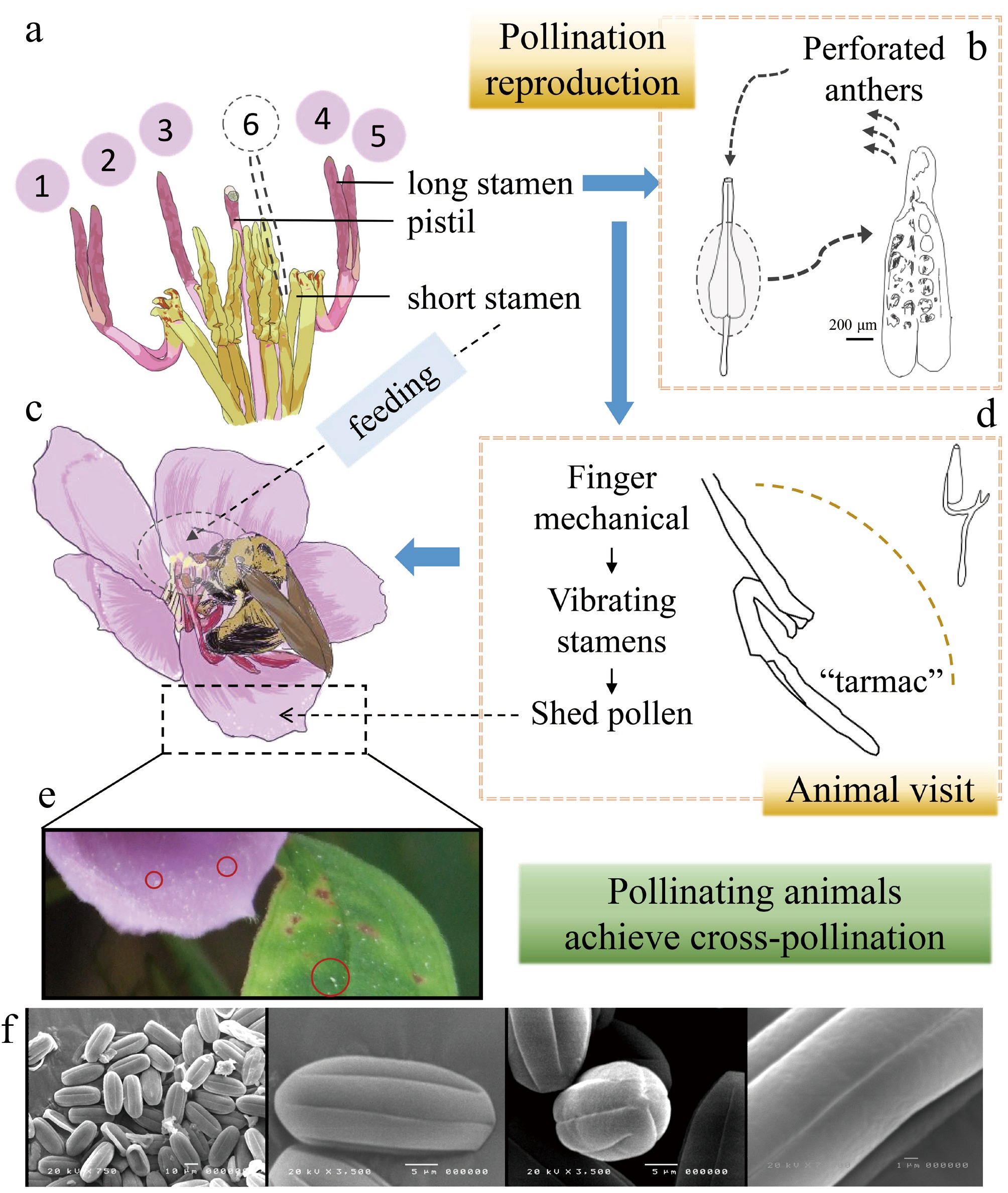
Figure 3.
Characteristics of heteromorphic stamens of Melastoma candidum and their co-evolution with pollinating animals. (a) Characteristics of heterosexual stamens. (b) The process of pollen release from long stamens. (c) The synergy between the heteromorphic stamen structure and the feeding behavior of Xylocopa aeratus. (d) The structure of long stamens. (e) After the Xylocopa aeratus feed, their pollen scatters on the flowers and leaves. (f) Electron microscope scanning image of M. candidum pollen[19].
-
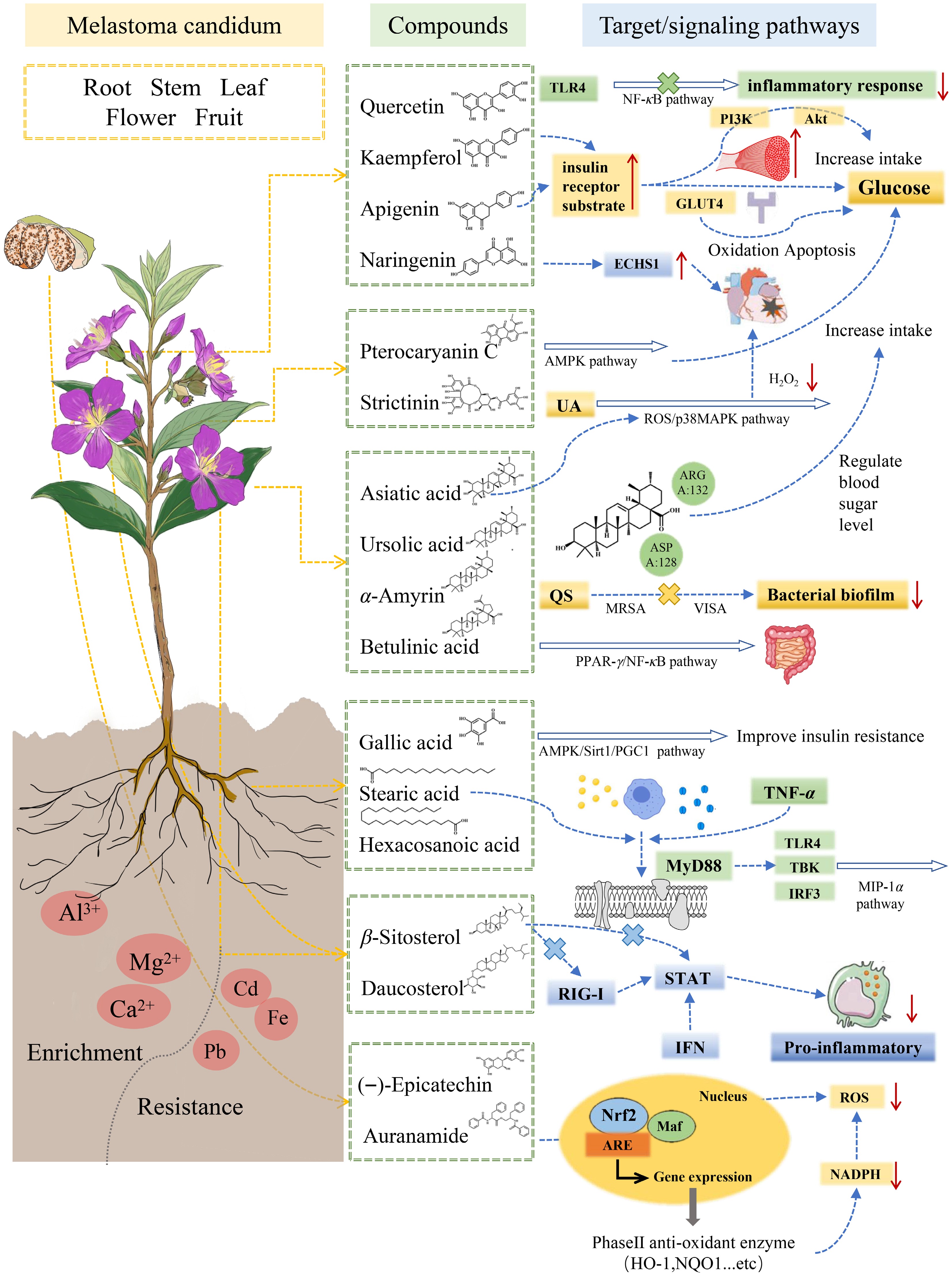
Figure 4.
Medicinal components of the tissues and organs of plants of Melastoma and their functional signaling pathways.
-
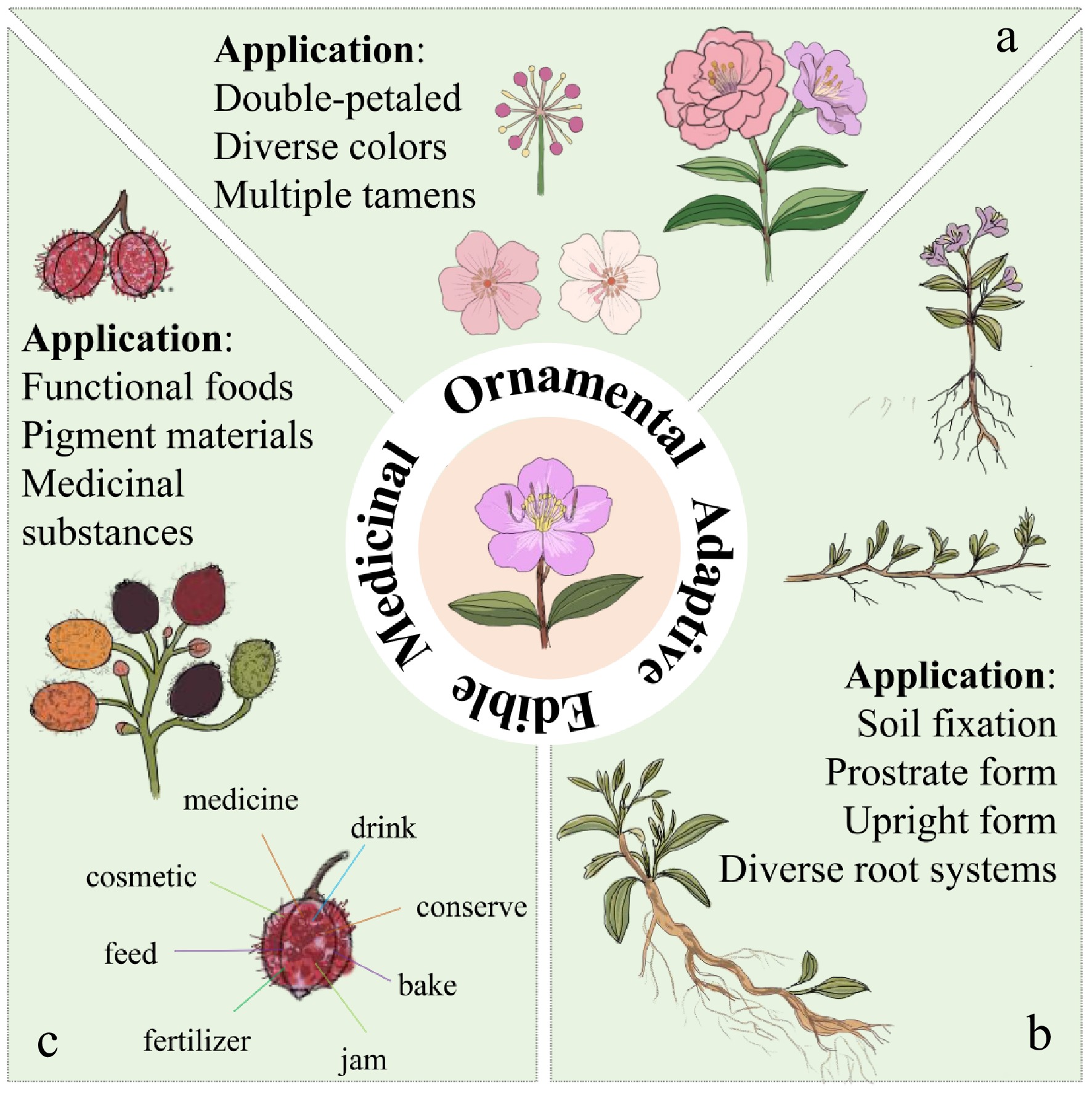
Figure 5.
Breeding directions of Melastoma. (a) In terms of ornamental applications, it includes the double-petal morphology of flowers, the diversity of flower colors, and the multiple stamens of heteromorphic stamens. (b) The aspects of adaptive application include the diversity of root systems and the upright/creeping forms of plants. (c) In terms of food and medicinal applications, it includes functional foods, fruit color, and medicinal substances such as alkaloids rich in fruits.
-
Types Representative compounds Chemical structural Target/signaling pathway Mechanism of action Species Ref. formula Flavonoids Quercetin 
The nf-kappa B Inhibition of the NF-κB pathway reduces TNF-α expression in LPS-induced RAW264.7 cells M. candidum
M. dodecandrum
M. malabathricum
M. normale
M. villosum[53, 59−62] Kaempferol 
PI3K/Akt/GLUT4 Up-regulate the gene and protein expression of PI3K, Akt, and GLUT4 in skeletal muscle, promote glucose transport and utilization, and reduce blood glucose M. candidum
M. dodecandrum
M. malabathricum
M. normale
M. villosum[55, 62,63] Naringenin 
ECHS1–PPARα/AMPK Reduces doxorubicin-induced myocardial oxidative stress and apoptosis by upregulating ECHS1 protein M. malabathricum [64,65] Apigenin 
PI3K/Akt/GLUT4 Up-regulated the expression of GLUT4 and improved glucose consumption and glycogen synthesis in IR-HepG2 cells. Slightly increased expression of PI3K and p-Akt M. candidum
M. dodecandrum
M. candidum[63, 66] Tannins Strictinin 
ROS/p38MAPK Provide hydrogen atoms to remove ROS and reduce oxidative stress; UA regulates the p38MAPK pathway and inhibits H2O2-induced apoptosis M. malabathricum
M. dodecandrum
M. normale[67−69] Terpenoids Asiatic acid 
MAPK/STAT3 Activation of ERK and p38 MAPK pathways induces apoptosis and cell cycle arrest in breast cancer cells; Inhibition of STAT3 and Claudin-1 M. dodecandrum [65, 70] Ursolic acid 
AMPK /PPARα Hypoglycemic mechanisms M. malabathricum
M. dodecandrum
M. intermedium[61, 71] Betulinic acid 
PPAR-γ/NF-κB Improvement of intestinal inflammation through the PPAR-γ/NF-κB pathway M. malabathricum
M. dodecandrum
M. intermedium[68, 72] Organic acids Gallic acid 
AMPK/Sirt1/PGC1 Promotes mitochondrial function and improves insulin resistance M. candidum
M. intermedium
M. malabathricum
M. polyanthum
M. normale
M. affine[73,74] Stearic acid 
TLR4/TBK/IRF3 Enhance MIP-1α expression and promote inflammatory response; Activates the lactate-HIF1α pathway at high concentrations, upregulating VEGF and pro-inflammatory cytokines M. dodecandrum [62, 75] Others (-)-Epicatechin 
Nrf2 Promote the expression of antioxidant enzymes (HO-1, NQO1), inhibit NADPH oxidase activity, and reduce ROS production M. dodecandrum [65, 73] Auranamide 
NF-κB/Nrf2 Provide hydrogen atoms to remove ROS and reduce oxidative stress; UA regulates the p38MAPK pathway and inhibits H2O2-induced apoptosis M. malabathricum [75] Table 1.
Types, representative compounds, and mechanisms of action of bioactive compounds in the genus Melastoma.
-
Species Genome size
(Mb)Anchored
pseudo-chromosomesScaffold/contig N50 (Mb) Repetitive sequence Protein-coding genes Functional annotation (%) ncRNAs M. candidum 256.2 12 20.5 (scaffold) 31.5% 40,938 91.3 1,818 M. dodecandrum 299.81 12 3.00 (contig) 40.9% 35,681 98.98 1,818 (miRNA 105, tRNA 633) Table 2.
The nuclear genome of Melastoma candidum and Melastoma dodecandrum.
-
Species Genome size
(bp)GC content Structure Repetitive
sequenceM. dodecandrum 411,944 44.18% Circular 3.52% M. candidum 391,595 44.36% Circular 3.52%–3.81% M. sanguineum 395,542 44.37% Circular 3.52%–3.81% IDT events exist in the mitochondrial genomes of eight species, reflecting a common ancestral origin. Table 3.
The mitochondrial genome of M. dodecandrum, M. candidum, and M. sanguineum.
-
Species GenBank accession number Plastome length (bp) LSC (bp) SSC (bp) IR (bp) GC content (%) Allomaieta villosa KX826819 156,452 85,914 16,975 26,781 36.9 Bertolonia acuminata KX826820 156,045 85,571 17,011 26,733 37.0 Blakea schlimii KX826821 155,862 85,370 16,998 26,747 37.1 Eriocnema fulva KX826822 155,994 85,431 16,953 26,805 37.0 Graffenrieda moritziana KX826823 155,733 85,341 16,924 26,734 37.0 Henriettea barkeri KX826824 156,527 85,991 17,036 26,750 36.9 Merianthera pulchra KX826825 156,168 85,621 17,001 26,773 37.0 Miconia dodecandra KX826826 157,216 86,609 16,999 26,804 37.0 Nepsera aquatica KX826827 155,110 84,644 17,066 26,700 37.1 Opisthocentra clidemioide KX826828 156,352 85,866 16,942 26,772 37.0 Pterogastra divaricata KX826829 154,948 84,718 17,156 26,537 37.2 Rhexia virginica KX826830 154,635 84,459 16,924 26,626 37.2 Rhynchanthera bracteata KX826831 155,108 85,093 16,729 26,643 37.0 Salpinga maranoniensi KX826832 153,311 85,128 16,653 25,765 37.4 Tibouchina longifolia KX826833 156,789 86,297 17,124 26,684 37.1 Triolena amazonica KX826834 156,652 86,200 16,970 26,741 36.9 Melastoma candidum KY745894 156,682 86,084 17,094 26,752 37.17 Tigridiopalma magnifica MF663760 155,663 85,161 16,932 26,785 37.11 Melastoma dodecandrum MH748092 156,611 86,014 17,097 26,750 37.1 Scorpiothyrsus erythrotrichus MZ434958 160,731 85,482 17,007 26,780 36.9 Table 4.
The chloroplast genome of melastomataceae.
Figures
(5)
Tables
(4)