-
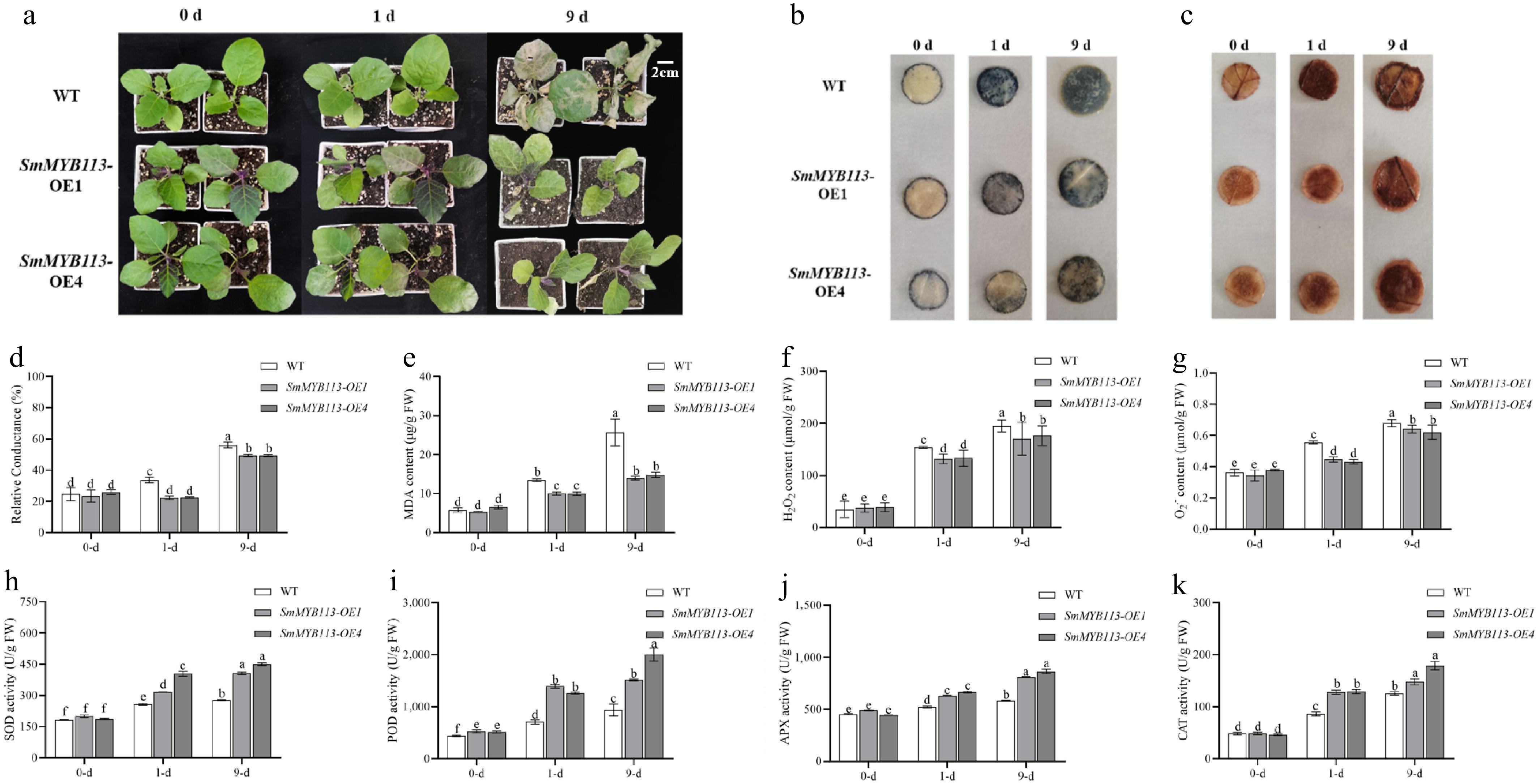
Figure 1.
Phenotypic and physiological differences between WT and SmMYB113-OE plants under low-temperature stress. (a) Phenotypic differences under 5 °C (16 h light/8 h dark) at 0, 1, and 9 d. (b) DAB staining for H2O2 localization. (c) NBT staining for O2·− localization. (d) Relative electrolyte leakage (REL). (e) MDA content. (f) Colorimetric quantification of H2O2 content. (g) Hydroxylamine oxidation assay for O2·− content. (h)–(k) Activities of antioxidant enzymes SOD, POD, APX, and CAT. Values represent means ± SD of three independent biological replicates (n = 3). Significant differences were determined by one-way ANOVA followed by Tukey's test (p < 0.05). Different lowercase letters indicate significant differences (p < 0.05).
-
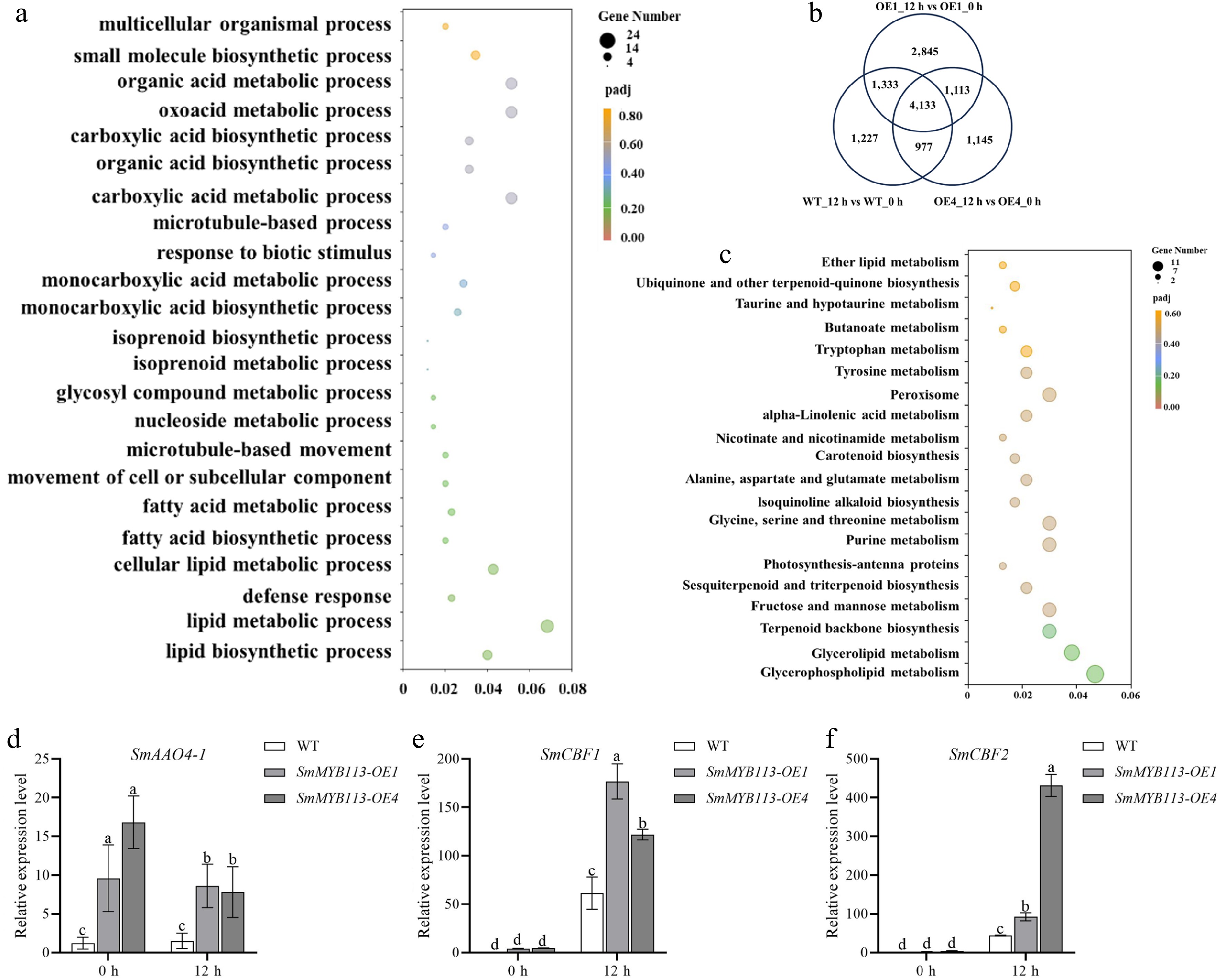
Figure 2.
Transcriptomic analysis reveals synergistic cold response pathways. (a) Gene Ontology (GO) enrichment analysis of the 1,113 DEGs common to OE lines but absent in WT. (b) Venn diagram of DEGs identified in WT, OE1, and OE4 plants under cold stress. (c) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the 1,113 DEGs. (d)–(f) Expression levels of key genes in OE and WT plants under 5 °C (16 h light/8 h dark) at 0 and 12 h: (d) ABA biosynthetic gene SmAAO4-1, and cold response factors (e) SmCBF1, and (f) SmCBF2. Values represent means ± SD of three independent biological replicates (n = 3). Significant differences were determined by one-way ANOVA followed by Tukey's test (p < 0.05). Different lowercase letters indicate significant differences (p < 0.05).
-
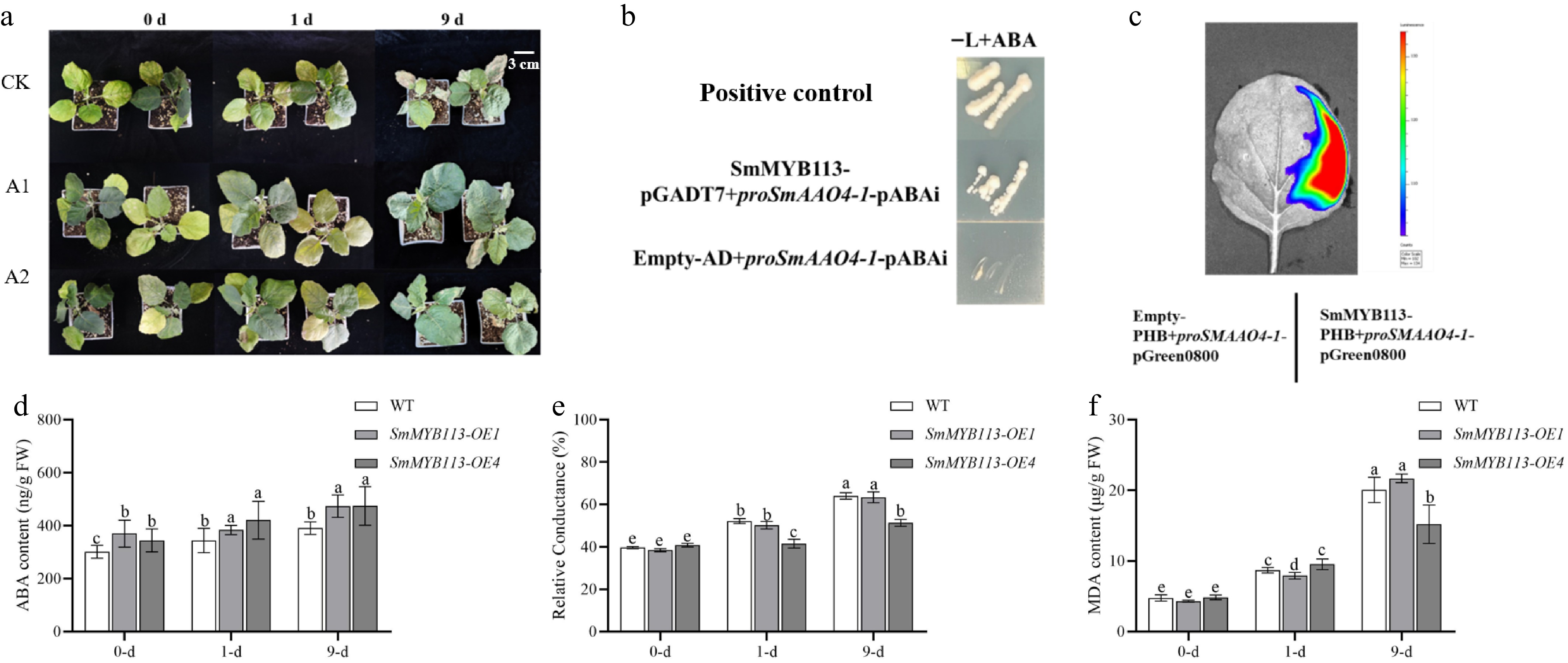
Figure 3.
Role of ABA in cold tolerance and regulation of SmAAO4-1 by SmMYB113. (a) Phenotype of WT plants under 5 °C (16 h light/8 h dark) at 0, 1, and 9 d after pretreatment: CK (no pretreatment), A1 (water spray), A2 (ABA spray). (b) Y1H assay showing interaction of SmMYB113 with the SmAAO4-1 promoter. Yeast growth on SD/-Leu medium with AbA (100 ng·mL−1). (c) LCI assay in tobacco leaves confirming SmMYB113 binding to the SmAAO4-1 promoter and activating luciferase expression. Physiological indices in WT, OE1, and OE4 leaves under 5 °C (16 h light/8 h dark) at 0, 1, and 9 d: (d) ABA content, (e) Relative electrolyte leakage (REL), (f) MDA content. Values represent means ± SD of three independent biological replicates (n = 3). Significant differences were determined by one-way ANOVA followed by Tukey's test (p < 0.05). Different lowercase letters indicate significant differences (p < 0.05).
-
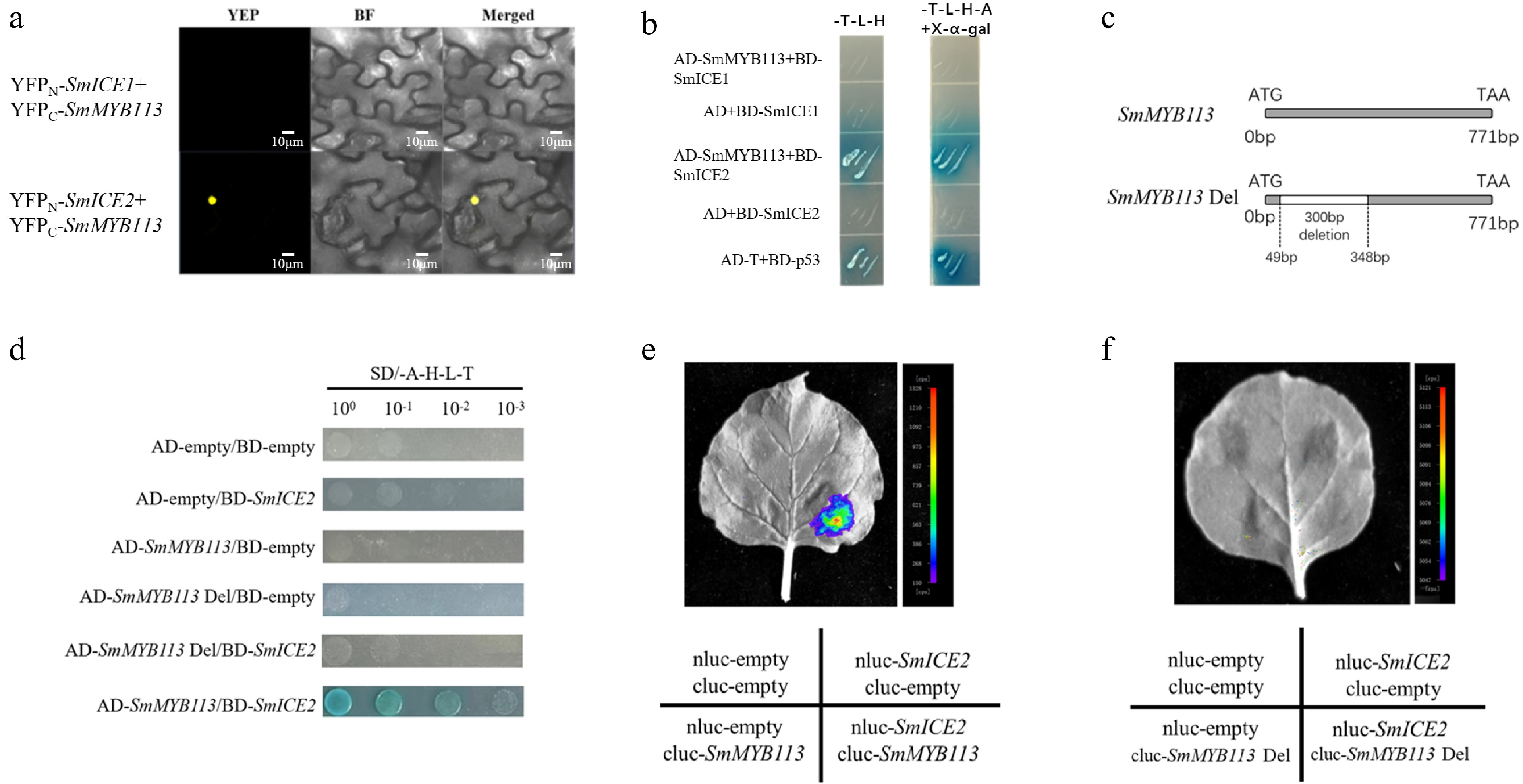
Figure 4.
Validation of SmMYB113-SmICE2 interaction and identification of the interaction domain. (a) BiFC assay showing SmICE2-SmMYB113 interaction in tobacco leaf nuclei. SmICE1/YFPN and SmICE2/YFPN were paired with SmMYB113/YFPC. (b) Y2H assay confirming SmICE2-SmMYB113 interaction on SD/-T/-L/-H/-A medium. (c) Schematic diagram of SmMYB113 deletion mutant (SmMYB113Del, lacking residues 49−348). (d) Y2H assay showing loss of interaction between SmMYB113Del and SmICE2. (e) LCI assay showing strong fluorescence signal upon co-expression of full-length SmMYB113 and SmICE2. (f) LCI assay showing loss of fluorescence signal upon co-expression of SmMYB113Del and SmICE2.
-
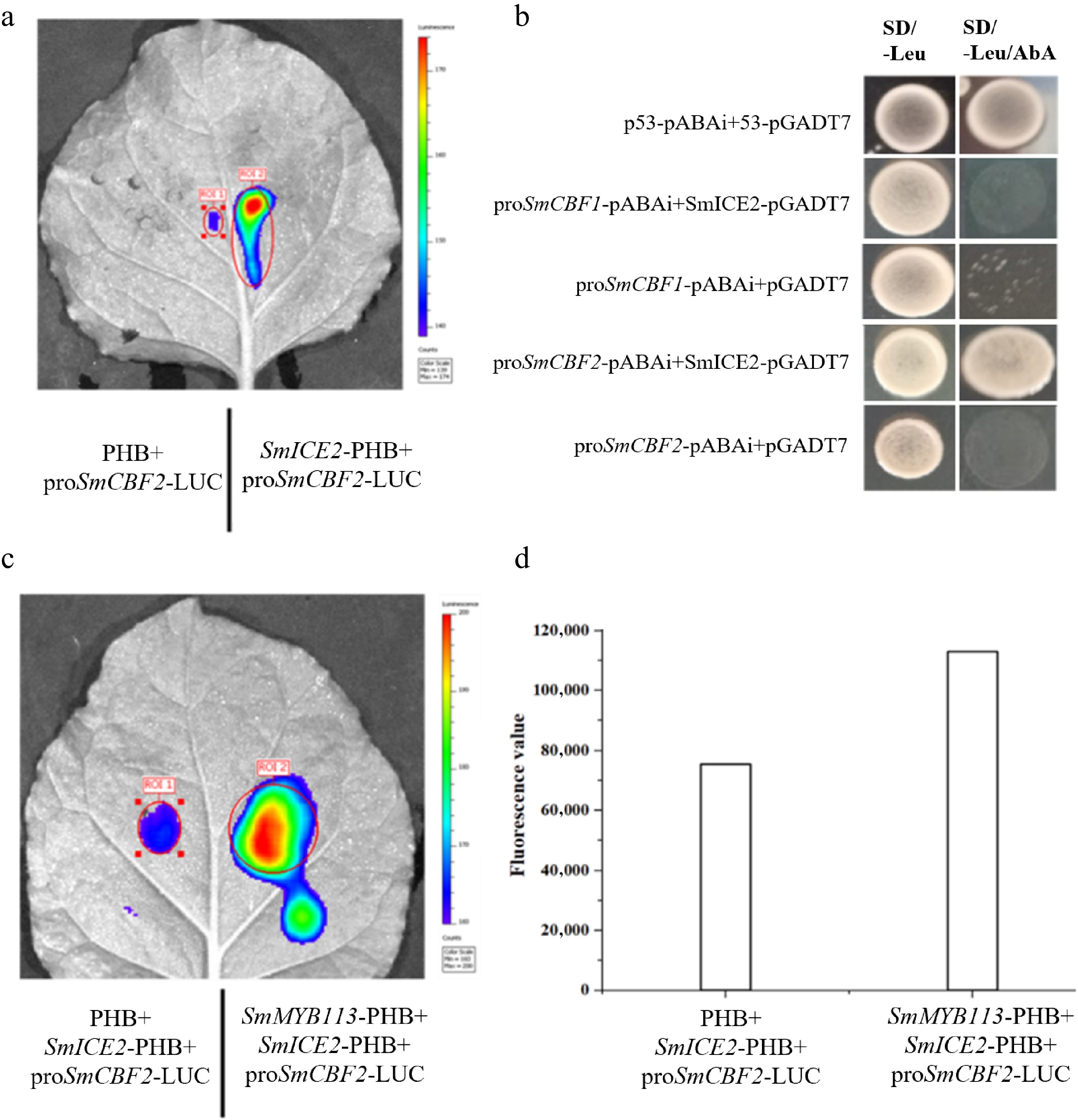
Figure 5.
SmMYB113-SmICE2 interaction enhances SmICE2-mediated transcriptional activation of SmCBF2. (a) LCI assay showing SmICE2 binding to and activating the SmCBF2 promoter. (b) Y1H assay confirming SmICE2 activation of SmCBF2 expression. (c) LCI assay demonstrating that co-expression of SmMYB113 and SmICE2 synergistically enhances luciferase activity driven by the SmCBF2 promoter compared to SmICE2 alone. (d) Quantification of luminescence intensity from (c).
-
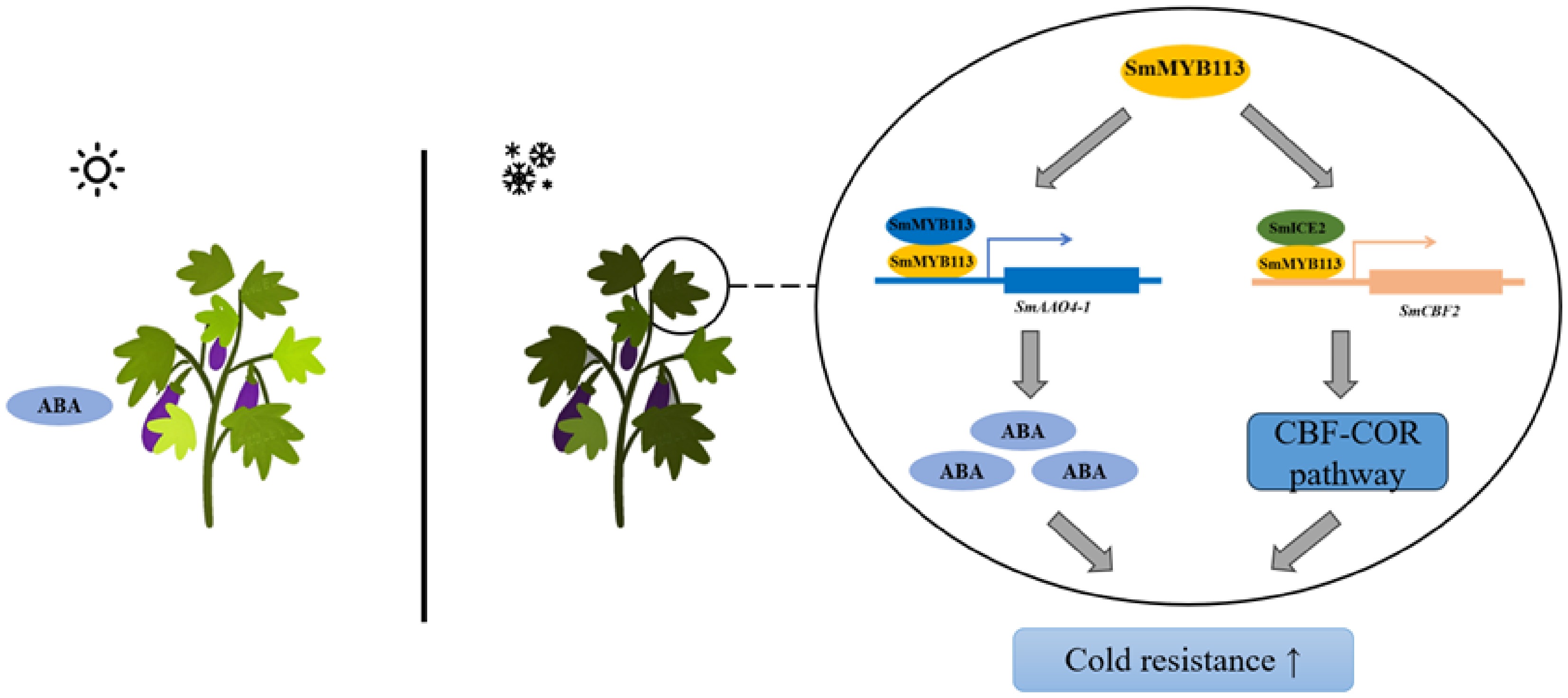
Figure 6.
Proposed model of SmMYB113-mediated cold tolerance in eggplant. Upon exposure to low temperature, SmMYB113 enhances cold tolerance through two mechanisms: (1) Transcriptional activation of SmAAO4-1, promoting ABA biosynthesis; (2) Physical interaction with SmICE2, enhancing SmICE2's transcriptional activation of SmCBF2, thereby participating in the ICE-CBF pathway.
Figures
(6)
Tables
(0)